Aldehyde-induced DNA-protein crosslinks- DNA damage, repair and mutagenesis
- PMID: 39328207
- PMCID: PMC11424613
- DOI: 10.3389/fonc.2024.1478373
Aldehyde-induced DNA-protein crosslinks- DNA damage, repair and mutagenesis
Abstract
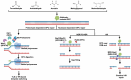
Aldehyde exposure has been shown to lead to the formation of DNA damage comprising of DNA-protein crosslinks (DPCs), base adducts and interstrand or intrastrand crosslinks. DPCs have recently drawn more attention because of recent advances in detection and quantification of these adducts. DPCs are highly deleterious to genome stability and have been shown to block replication forks, leading to wide-spread mutagenesis. Cellular mechanisms to prevent DPC-induced damage include excision repair pathways, homologous recombination, and specialized proteases involved in cleaving the covalently bound proteins from DNA. These pathways were first discovered in formaldehyde-treated cells, however, since then, various other aldehydes have been shown to induce formation of DPCs in cells. Defects in DPC repair or aldehyde clearance mechanisms lead to various diseases including Ruijs-Aalfs syndrome and AMeD syndrome in humans. Here, we discuss recent developments in understanding how aldehydes form DPCs, how they are repaired, and the consequences of defects in these repair pathways.
Keywords: DNA damage; DNA protein crosslink; DNA repair; aldehydes; genome instability.
Copyright © 2024 Blouin and Saini.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Mechanisms and Regulation of DNA-Protein Crosslink Repair During DNA Replication by SPRTN Protease.Front Mol Biosci. 2022 Jun 15;9:916697. doi: 10.3389/fmolb.2022.916697. eCollection 2022. Front Mol Biosci. 2022. PMID: 35782873 Free PMC article. Review.
-
Endogenous aldehyde-induced DNA-protein crosslinks are resolved by transcription-coupled repair.Nat Cell Biol. 2024 May;26(5):784-796. doi: 10.1038/s41556-024-01401-2. Epub 2024 Apr 10. Nat Cell Biol. 2024. PMID: 38600234 Free PMC article.
-
Metalloprotease SPRTN/DVC1 Orchestrates Replication-Coupled DNA-Protein Crosslink Repair.Mol Cell. 2016 Nov 17;64(4):704-719. doi: 10.1016/j.molcel.2016.09.032. Epub 2016 Oct 27. Mol Cell. 2016. PMID: 27871366 Free PMC article.
-
A quantitative PCR-based assay reveals that nucleotide excision repair plays a predominant role in the removal of DNA-protein crosslinks from plasmids transfected into mammalian cells.DNA Repair (Amst). 2018 Feb;62:18-27. doi: 10.1016/j.dnarep.2018.01.004. Epub 2018 Jan 9. DNA Repair (Amst). 2018. PMID: 29413806 Free PMC article.
-
Formation and repair of DNA-protein crosslink damage.Sci China Life Sci. 2017 Oct;60(10):1065-1076. doi: 10.1007/s11427-017-9183-4. Epub 2017 Oct 30. Sci China Life Sci. 2017. PMID: 29098631 Free PMC article. Review.
Cited by
-
Role of Redox-Induced Protein Modifications in Spermatozoa in Health and Disease.Antioxidants (Basel). 2025 Jun 12;14(6):720. doi: 10.3390/antiox14060720. Antioxidants (Basel). 2025. PMID: 40563353 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

