Prognostic importance of splicing-triggered aberrations of protein complex interfaces in cancer
- PMID: 39328266
- PMCID: PMC11426328
- DOI: 10.1093/nargab/lqae133
Prognostic importance of splicing-triggered aberrations of protein complex interfaces in cancer
Abstract
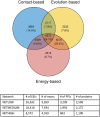
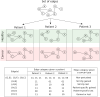
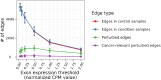
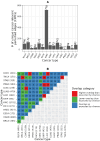
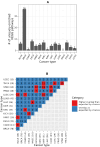
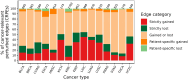
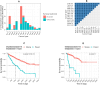
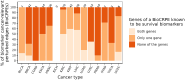
Aberrant alternative splicing (AS) is a prominent hallmark of cancer. AS can perturb protein-protein interactions (PPIs) by adding or removing interface regions encoded by individual exons. Identifying prognostic exon-exon interactions (EEIs) from PPI interfaces can help discover AS-affected cancer-driving PPIs that can serve as potential drug targets. Here, we assessed the prognostic significance of EEIs across 15 cancer types by integrating RNA-seq data with three-dimensional (3D) structures of protein complexes. By analyzing the resulting EEI network we identified patient-specific perturbed EEIs (i.e., EEIs present in healthy samples but absent from the paired cancer samples or vice versa) that were significantly associated with survival. We provide the first evidence that EEIs can be used as prognostic biomarkers for cancer patient survival. Our findings provide mechanistic insights into AS-affected PPI interfaces. Given the ongoing expansion of available RNA-seq data and the number of 3D structurally-resolved (or confidently predicted) protein complexes, our computational framework will help accelerate the discovery of clinically important cancer-promoting AS events.
© The Author(s) 2024. Published by Oxford University Press on behalf of NAR Genomics and Bioinformatics.
Figures


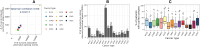

References
LinkOut - more resources
Full Text Sources
Research Materials

