Comparing NGS-Based identification of bloodstream infections to traditional culture methods for enhanced ICU care: a comprehensive study
- PMID: 39328359
- PMCID: PMC11424606
- DOI: 10.3389/fcimb.2024.1454549
Comparing NGS-Based identification of bloodstream infections to traditional culture methods for enhanced ICU care: a comprehensive study
Erratum in
-
Corrigendum: Comparing NGS-based identification of bloodstream infections to traditional culture methods for enhanced ICU care: a comprehensive study.Front Cell Infect Microbiol. 2025 Mar 27;15:1553275. doi: 10.3389/fcimb.2025.1553275. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40212848 Free PMC article.
Abstract
Background: Accurate identification of infectious diseases using molecular techniques, such as PCR and NGS, is well-established. This study aims to assess the utility of Bactfast and Fungifast in diagnosing bloodstream infections in ICU settings, comparing them against traditional culture methods. The objectives include evaluating sensitivity and specificity and identifying a wide range of pathogens, including non-culturable species.
Methods: We collected 500 non-duplicate blood samples from ICU patients between January 2023 and December 2023. Specimens underwent traditional culture, MALDI-TOF, VITEK®2 compact system, and NGS-based Bactfast and Fungifast analyses.
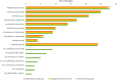
Results: Out of the 500 samples, 26.8% (n=134) showed bacterial growth via traditional culture methods, while 4.8% (n=24) were positive for fungal growth. MALDI-TOF and VITEK®2 compact system yielded comparable results, identifying 26.4% (n=132) of specimens with bacterial growth. NGS-based Bactfast detected bacterial presence in 38.2% (n=191) of samples, including non-culturable bacteria missed by traditional methods. However, NGS-based Fungifast showed concordant fungal detection rates with culture methods. Among identified pathogens by culture method included Klebsiella pneumoniae 20.89% (n=28), Enterococcus faecalis 18.65% (n=25), Escherichia coli 15.67% (n=21), Pseudomonas aeruginosa 12.68% (n=17), Acinetobacter baumannii 10.44% (n=14), various Streptococcus species 7.46% (n=10), Mycobacterium tuberculosis 6.71% (n=9), Mycobacterium abscessus 4.47% (n=6), and Salmonella spp 2.98% (n=4). Non-culture-based NGS identified additional (n=33) pathogens, including Klebsiella pneumoniae 27.27% (n=9), Bacteroides fragilis 21.21% (n=7), Aerococcus viridans 15.15% (n=5), Elizabethkingia anopheles 12.12% (n=4), Aeromonas salmonicida 9% (n=3), Clostridium 9% (n=3), and Bacteroides vulgatus 6% (n=2). Candida albicans was reported in 5% (n=24) of samples by both methods.
Conclusion: NGS-based Bactfast and Fungifast demonstrate high sensitivity in identifying a wide array of bacterial and fungal pathogens in ICU patients, outperforming traditional culture methods in detecting non-culturable organisms. These molecular assays offer rapid and comprehensive diagnostic capabilities, potentially improving clinical outcomes through timely and accurate pathogen identification.
Keywords: NGS-based identification in bloodstream infection next-generation sequencing (NGS); Bactfast and Fungifast; ICU pathogen identification; bloodstream infections (BSIs); molecular diagnostics.
Copyright © 2024 Wang, Chauhan, Luo, Sharma, Li and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
ICU care through NGS - Based identification of infectious agents: A comparative study.Heliyon. 2024 Jul 14;10(14):e34538. doi: 10.1016/j.heliyon.2024.e34538. eCollection 2024 Jul 30. Heliyon. 2024. PMID: 39082018 Free PMC article.
-
Molecular rapid diagnostic testing for bloodstream infections: Nanopore targeted sequencing with pathogen-specific primers.J Infect. 2024 Jun;88(6):106166. doi: 10.1016/j.jinf.2024.106166. Epub 2024 Apr 24. J Infect. 2024. PMID: 38670268
-
Rapid identification of bloodstream bacterial and fungal pathogens and their antibiotic resistance determinants from positively flagged blood cultures using the BioFire FilmArray blood culture identification panel.J Microbiol Immunol Infect. 2020 Dec;53(6):882-891. doi: 10.1016/j.jmii.2020.03.018. Epub 2020 Apr 2. J Microbiol Immunol Infect. 2020. PMID: 32305272
-
The Use of Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) for the Identification of Pathogens Causing Sepsis.J Appl Lab Med. 2019 Jan;3(4):675-685. doi: 10.1373/jalm.2018.027318. Epub 2018 Sep 13. J Appl Lab Med. 2019. PMID: 31639735 Review.
-
Metagenomics to Assist in the Diagnosis of Bloodstream Infection.J Appl Lab Med. 2019 Jan;3(4):643-653. doi: 10.1373/jalm.2018.026120. Epub 2018 Nov 30. J Appl Lab Med. 2019. PMID: 31639732 Review.
Cited by
-
Microbiota shifts in fracture-related infections and pathogenic transitions identified by 16S rDNA sequencing.Sci Rep. 2025 Mar 5;15(1):7732. doi: 10.1038/s41598-025-91990-1. Sci Rep. 2025. PMID: 40044740 Free PMC article.
-
Development of a Species-Specific PCR Assay for Aerococcus urinaeequi Using Whole Genome Sequencing.Pathogens. 2025 Jun 25;14(7):634. doi: 10.3390/pathogens14070634. Pathogens. 2025. PMID: 40732682 Free PMC article.
-
Healthcare-Associated Infections: The Role of Microbial and Environmental Factors in Infection Control-A Narrative Review.Infect Dis Ther. 2025 May;14(5):933-971. doi: 10.1007/s40121-025-01143-0. Epub 2025 Apr 10. Infect Dis Ther. 2025. PMID: 40208412 Free PMC article. Review.
References
-
- Anahtar M. N., Shaw B., Slater D., Byrne E., Botti-Lodovico Y., Adams G., et al. . (2020). Development of a qualitative real-time RT-PCR assay for the detection of SARS-CoV-2: A guide and case study in setting up an emergency-use, laboratory-developed molecular assay. medRxiv 2020, 8.26.20157297. doi: 10.1101/2020.08.26.20157297 - DOI - PMC - PubMed
-
- BactFast Micro genomics India. Available online at: https://www.microgenomics.in/bactfast/ (Accessed June 18, 2024).
-
- Blot S., Ruppé E., Harbarth S., Asehnoune K., Poulakou G., Luyt C.-E., et al. . (2022). Healthcare-associated infections in adult intensive care unit patients: Changes in epidemiology, diagnosis, prevention and contributions of new technologies. Intensive Crit. Care Nurs. 70, 103227. doi: 10.1016/j.iccn.2022.103227 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous