Integrated single-cell and bulk RNA sequencing analysis reveal immune-related biomarkers in postmenopausal osteoporosis
- PMID: 39328516
- PMCID: PMC11425179
- DOI: 10.1016/j.heliyon.2024.e38022
Integrated single-cell and bulk RNA sequencing analysis reveal immune-related biomarkers in postmenopausal osteoporosis
Abstract
Background: Postmenopausal osteoporosis (PMOP) represents as a significant health concern, particularly as the population ages. Currently, there is a paucity of comprehensive descriptions regarding the immunoregulatory mechanisms and early diagnostic biomarkers associated with PMOP. This study aims to examine immune-related differentially expressed genes (IR-DEGs) in the peripheral blood mononuclear cells of PMOP patients to identify immunological patterns and diagnostic biomarkers.
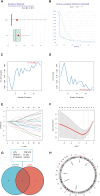
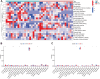
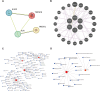
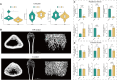
Methods: The GSE56815 dataset from the Gene Expression Omnibus (GEO) database was used as the training group, while the GSE2208 dataset served as the validation group. Initially, differential expression analysis was conducted after data integration to identify IR-DEGs in the peripheral blood mononuclear cells of PMOP. Subsequently, feature selection of these IR-DEGs was performed using RF, SVM-RFE, and LASSO regression models. Additionally, the expression of IR-DEGs in distinct bone marrow cell subtypes was analyzed using single-cell RNA sequencing (scRNA-seq) datasets, allowing the identification of cellular communication patterns within various cell subgroups. Finally, molecular subtypes and diagnostic models for PMOP were constructed based on these selected IR-DEGs. Furthermore, the expression levels of characteristic IR-DEGs were examined in rat osteoporosis (OP) models.
Results: Using machine learning, six IR-DEGs (JUN, HMOX1, CYSLTR2, TNFSF8, IL1R2, and SSTR5) were identified. Subsequently, two molecular subtypes of PMOP (subtype 1 and subtype 2) were established, with subtype 1 exhibiting a higher proportion of M1 macrophage infiltration. Analysis of the scRNA-seq dataset revealed 11 distinct cell clusters. It was noted that JUN was significantly overexpressed in M1 macrophages, while HMOX1 showed a marked elevation in endothelial cells and M2 macrophages. Cell communication results suggested that the PMOP microenvironment features increased interactions among M2 macrophages, CD8+ T cells, Tregs, and fibroblasts. The diagnostic model based on these six IR-DEGs demonstrated excellent diagnostic performance (AUC = 0.927). In the OP rat model, the expression of IL1R2 and TNFSF8 were significantly elevated.
Conclusion: JUN, HMOX1, CYSLTR2, TNFSF8, IL1R2, and SSTR5 may serve as promising molecular targets for diagnosing and subtyping patients with PMOP. These results offer novel perspectives on the early diagnosis of PMOP and the advancement of personalized immune-based therapies.
Keywords: Biomarkers; Diagnosis; Immune; Molecular subtype; Postmenopausal osteoporosis.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:Haidong Li reports article publishing charges was provided by Natural Science Foundation of Zhejiang Province. If there are other authors, they declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures








References
-
- Black D.M., Rosen C.J. Clinical practice. Postmenopausal osteoporosis. N. Engl. J. Med. 2016;374(3):254–262. - PubMed
-
- Recker R., Lappe J., Davies K., Heaney R. Characterization of perimenopausal bone loss: a prospective study. J. Bone Miner. Res. 2000;15(10):1965–1973. - PubMed
-
- Looker A.C., Orwoll E.S., Johnston C.C., Jr., Lindsay R.L., Wahner H.W., Dunn W.L., et al. Prevalence of low femoral bone density in older U.S. adults from NHANES III. J. Bone Miner. Res. 1997;12(11):1761–1768. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

