Aggregation Rules of Short Peptides
- PMID: 39328768
- PMCID: PMC11423302
- DOI: 10.1021/jacsau.4c00501
Aggregation Rules of Short Peptides
Abstract
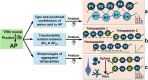
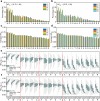
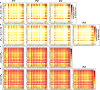
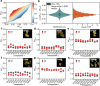
The elucidation of aggregation rules for short peptides (e.g., tetrapeptides and pentapeptides) is crucial for the precise manipulation of aggregation. In this study, we derive comprehensive aggregation rules for tetrapeptides and pentapeptides across the entire sequence space based on the aggregation propensity values predicted by a transformer-based deep learning model. Our analysis focuses on three quantitative aspects. First, we investigate the type and positional effects of amino acids on aggregation, considering both the first- and second-order contributions. By identifying specific amino acids and amino acid pairs that promote or attenuate aggregation, we gain insights into the underlying aggregation mechanisms. Second, we explore the transferability of aggregation propensities between tetrapeptides and pentapeptides, aiming to explore the possibility of enhancing or mitigating aggregation by concatenating or removing specific amino acids at the termini. Finally, we evaluate the aggregation morphologies of over 20,000 tetrapeptides, regarding the morphology distribution and type and positional contributions of each amino acid. This work extends the existing aggregation rules from tripeptide sequences to millions of tetrapeptide and pentapeptide sequences, offering experimentalists an explicit roadmap for fine-tuning the aggregation behavior of short peptides for diverse applications, including hydrogels, emulsions, or pharmaceuticals.
© 2024 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
Deep Learning Empowers the Discovery of Self-Assembling Peptides with Over 10 Trillion Sequences.Adv Sci (Weinh). 2023 Nov;10(31):e2301544. doi: 10.1002/advs.202301544. Epub 2023 Sep 25. Adv Sci (Weinh). 2023. PMID: 37749875 Free PMC article.
-
Complexes of technetium-99m with tetrapeptides, a new class of 99mTc-labelled agents.Nucl Med Biol. 1995 Apr;22(3):325-38. doi: 10.1016/0969-8051(94)00110-6. Nucl Med Biol. 1995. PMID: 7627148
-
Opioid receptor affinity and selectivity effects of second residue and carboxy terminal residue variation in a cyclic disulfide-containing opioid tetrapeptide.Int J Pept Protein Res. 1991 Mar;37(3):224-9. doi: 10.1111/j.1399-3011.1991.tb00274.x. Int J Pept Protein Res. 1991. PMID: 1651290
-
[A turning point in the knowledge of the structure-function-activity relations of elastin].J Soc Biol. 2001;195(2):181-93. J Soc Biol. 2001. PMID: 11727705 Review. French.
-
Dietary glycation compounds - implications for human health.Crit Rev Toxicol. 2024 Sep;54(8):485-617. doi: 10.1080/10408444.2024.2362985. Epub 2024 Aug 16. Crit Rev Toxicol. 2024. PMID: 39150724
Cited by
-
Interplay of Hydrophobicity, Charge, and Sequence Length in Oligopeptide Coassembly.J Phys Chem B. 2025 May 8;129(18):4383-4391. doi: 10.1021/acs.jpcb.5c00737. Epub 2025 Apr 23. J Phys Chem B. 2025. PMID: 40267030
References
LinkOut - more resources
Full Text Sources
