Whole-genome comparison using complete genomes from Campylobacter fetus strains revealed single nucleotide polymorphisms on non-genomic islands for subspecies differentiation
- PMID: 39328909
- PMCID: PMC11424552
- DOI: 10.3389/fmicb.2024.1452564
Whole-genome comparison using complete genomes from Campylobacter fetus strains revealed single nucleotide polymorphisms on non-genomic islands for subspecies differentiation
Abstract
Introduction: Bovine Genital Campylobacteriosis (BGC), caused by Campylobacter fetus subsp. venerealis, is a sexually transmitted bacterium that significantly impacts cattle reproductive performance. However, current detection methods lack consistency and reliability due to the close genetic similarity between C. fetus subsp. venerealis and C. fetus subsp. fetus. Therefore, this study aimed to utilize complete genome analysis to distinguish genetic features between C. fetus subsp. venerealis and other subspecies, thereby enhancing BGC detection for routine screening and epidemiological studies.
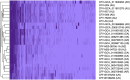
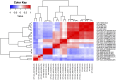
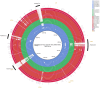
Methods and results: This study reported the complete genomes of four C. fetus subsp. fetus and five C. fetus subsp. venerealis, sequenced using long-read sequencing technologies. Comparative whole-genome analyses (n = 25) were conducted, incorporating an additional 16 complete C. fetus genomes from the NCBI database, to investigate the genomic differences between these two closely related C. fetus subspecies. Pan-genomic analyses revealed a core genome consisting of 1,561 genes and an accessory pangenome of 1,064 genes between the two C. fetus subspecies. However, no unique predicted genes were identified in either subspecies. Nonetheless, whole-genome single nucleotide polymorphisms (SNPs) analysis identified 289 SNPs unique to one or the C. fetus subspecies. After the removal of SNPs located on putative genomic islands, recombination sites, and those causing synonymous amino acid changes, the remaining 184 SNPs were functionally annotated. Candidate SNPs that were annotated with the KEGG "Peptidoglycan Biosynthesis" pathway were recruited for further analysis due to their potential association with the glycine intolerance characteristic of C. fetus subsp. venerealis and its biovar variant. Verification with 58 annotated C. fetus genomes, both complete and incomplete, from RefSeq, successfully classified these seven SNPs into two groups, aligning with their phenotypic identification as CFF (Campylobacter fetus subsp. fetus) or CFV/CFVi (Campylobacter fetus subsp. venerealis and its biovar variant). Furthermore, we demonstrated the application of mraY SNPs for detecting C. fetus subspecies using a quantitative PCR assay.
Discussion: Our results highlighted the high genetic stability of C. fetus subspecies. Nevertheless, Campylobacter fetus subsp. venerealis and its biovar variants encoded common SNPs in genes related to glycine intolerance, which differentiates them from C. fetus subsp. fetus. This discovery highlights the potential of employing a multiple-SNP assay for the precise differentiation of C. fetus subspecies.
Keywords: Campylobacter fetus; SNPs; complete genomes; genome comparison; glycine; subspecies; veterinary science.
Copyright © 2024 Ong, Blackall, Boe-Hansen, deWet, Hayes, Indjein, Korolik, Minchin, Nguyen, Nordin, Siddle, Turni, Venus, Westman, Zhang and Tabor.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures



Similar articles
-
Genomic analysis of Campylobacter fetus subspecies: identification of candidate virulence determinants and diagnostic assay targets.BMC Microbiol. 2009 May 8;9:86. doi: 10.1186/1471-2180-9-86. BMC Microbiol. 2009. PMID: 19422718 Free PMC article.
-
Campylobacter fetus subspecies specific PCR assays inferred from comparative genomic analysis for accurate subspecies identification.J Microbiol Methods. 2024 Nov;226:107049. doi: 10.1016/j.mimet.2024.107049. Epub 2024 Sep 27. J Microbiol Methods. 2024. PMID: 39343039
-
Comparative pangenomic analysis of Campylobacter fetus isolated from Spanish bulls and other mammalian species.Sci Rep. 2024 Feb 22;14(1):4347. doi: 10.1038/s41598-024-54750-1. Sci Rep. 2024. PMID: 38388650 Free PMC article.
-
So close and yet so far - Molecular Microbiology of Campylobacter fetus subspecies.Eur J Microbiol Immunol (Bp). 2012 Mar;2(1):66-75. doi: 10.1556/EuJMI.2.2012.1.10. Epub 2012 Mar 17. Eur J Microbiol Immunol (Bp). 2012. PMID: 24611123 Free PMC article. Review.
-
The History of Bovine Genital Campylobacteriosis in the Face of Political Turmoil and Structural Change in Cattle Farming in Germany.Vet Sci. 2023 Nov 23;10(12):665. doi: 10.3390/vetsci10120665. Vet Sci. 2023. PMID: 38133216 Free PMC article. Review.
References
-
- Andrew S. (2010). FastQC: a Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed January 15, 2020).
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

