Architectural organization and in situ fusion protein structure of lymphocytic choriomeningitis virus
- PMID: 39329471
- PMCID: PMC11495036
- DOI: 10.1128/jvi.00640-24
Architectural organization and in situ fusion protein structure of lymphocytic choriomeningitis virus
Abstract
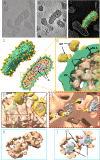
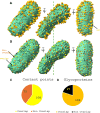
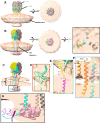
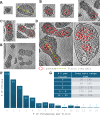
Arenaviruses exist globally and can cause hemorrhagic fever and neurological diseases, exemplified by the zoonotic pathogen lymphocytic choriomeningitis virus (LCMV). The structures of individual LCMV proteins or their fragments have been reported, but the architectural organization and the nucleocapsid assembly mechanism remain elusive. Importantly, the in situ structure of the arenavirus fusion protein complex (glycoprotein complex, GPC) as present on the virion prior to fusion, particularly with its integral stable signal peptide (SSP), has not been shown, hindering efforts such as structure-based vaccine design. Here, we have determined the in situ structure of LCMV proteins and their architectural organization in the virion by cryogenic electron tomography. The tomograms reveal the global distribution of GPC, matrix protein Z, and the contact points between the viral envelope and nucleocapsid. Subtomogram averaging yielded the in situ structure of the mature GPC with its transmembrane domain intact, revealing the GP2-SSP interface and the endodomain of GP2. The number of RNA-dependent RNA polymerase L molecules packaged within each virion varies, adding new perspectives to the infection mechanism. Together, these results delineate the structural organization of LCMV and offer new insights into its mechanism of LCMV maturation, egress, and cell entry.
Importance: The impact of COVID-19 on public health has highlighted the importance of understanding zoonotic pathogens. Lymphocytic choriomeningitis virus (LCMV) is a rodent-borne human pathogen that causes hemorrhagic fever. Herein, we describe the in situ structure of LCMV proteins and their architectural organization on the viral envelope and around the nucleocapsid. The virion structure reveals the distribution of the surface glycoprotein complex (GPC) and the contact points between the viral envelope and the underlying matrix protein, as well as the association with the nucleocapsid. The morphology and sizes of virions, as well as the number of RNA polymerase L inside each virion vary greatly, highlighting the fast-changing nature of LCMV. A comparison between the in situ GPC trimeric structure and prior ectodomain structures identifies the transmembrane and endo domains of GPC and key interactions among its subunits. The work provides new insights into LCMV assembly and informs future structure-guided vaccine design.
Keywords: arenavirus; cryogenic electron tomography; in situ structures; lymphocytic choriomeningitis virus; prefusion; spike proteins; virion.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Residues K465 and G467 within the Cytoplasmic Domain of GP2 Play a Critical Role in the Persistence of Lymphocytic Choriomeningitis Virus in Mice.J Virol. 2016 Oct 28;90(22):10102-10112. doi: 10.1128/JVI.01303-16. Print 2016 Nov 15. J Virol. 2016. PMID: 27581982 Free PMC article.
-
Crystal structure of the prefusion surface glycoprotein of the prototypic arenavirus LCMV.Nat Struct Mol Biol. 2016 Jun;23(6):513-521. doi: 10.1038/nsmb.3210. Epub 2016 Apr 25. Nat Struct Mol Biol. 2016. PMID: 27111888 Free PMC article.
-
Arenavirus stable signal peptide is the keystone subunit for glycoprotein complex organization.mBio. 2014 Oct 28;5(6):e02063. doi: 10.1128/mBio.02063-14. mBio. 2014. PMID: 25352624 Free PMC article.
-
Lymphocytic choriomeningitis virus: ways to establish and maintain non-cytolytic persistent infection.Acta Virol. 2016 Mar;60(1):15-26. doi: 10.4149/av_2016_01_15. Acta Virol. 2016. PMID: 26982463 Review.
-
Lymphocytic choriomeningitis virus: invisible but not innocent.Acta Virol. 2013;57(2):160-70. doi: 10.4149/av_2013_02_160. Acta Virol. 2013. PMID: 23600874 Review.
References
-
- Albariño CG, Palacios G, Khristova ML, Erickson BR, Carroll SA, Comer JA, Hui J, Briese T, St George K, Ksiazek TG, Lipkin WI, Nichol ST. 2010. High diversity and ancient common ancestry of lymphocytic choriomeningitis virus. Emerg Infect Dis 16:1093–1100. doi:10.3201/eid1607.091902 - DOI - PMC - PubMed

