Enormous diversity of RNA viruses in economic crustaceans
- PMID: 39329483
- PMCID: PMC11494968
- DOI: 10.1128/msystems.01016-24
Enormous diversity of RNA viruses in economic crustaceans
Abstract
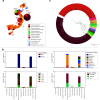
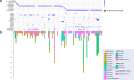
Crustaceans are important food sources worldwide and possess significant ecological status in the marine ecosystem. However, our understanding of the diversity and evolution of RNA viruses in crustaceans, especially in economic crustaceans, is still limited. Here, 106 batches of economic crustaceans including 13 species were collected from 24 locations in China during 2016-2021. We identified 90 RNA viruses, 69 of which were divergent from the known viruses. Viral transcripts were assigned to 18 different viral families/clades and three unclassified groups. Among the identified viruses, five were double-stranded RNA viruses, 74 were positive-sense single-stranded RNA (+ssRNA) viruses, nine were negative-sense single-stranded RNA (-ssRNA) viruses, and two belonged to an unclassified RNA virus group. Phylogenetic analyses showed that crustacean viruses were often clustered with viruses identified from invertebrates. Remarkably, most crustacean viruses were closely related to those from different host species along the same food chain or ecological aquatic niche. In addition, the genome structures of the newly discovered picornaviruses exhibited remarkable diversity. Our study significantly expands the diversity of viruses in important economic crustaceans and provides essential data for the risk assessment of the pathogens spreading in the global aquaculture industry.
Importance: The study delves into the largely uncharted territory of RNA viruses in crustaceans, which are not only vital for global food supply but also play a pivotal role in marine ecosystems. Focusing on economic crustaceans, the research uncovers 90 RNA viruses, with 69 being potentially new to science, highlighting the vast unknown viral diversity within these marine organisms. The findings reveal that these viruses are often related to those found in other invertebrates and tend to share close relationships with viruses from species within the same food web or habitat. This suggests that viruses may move between different marine species more frequently than previously thought. The discovery of such a wide variety of viruses, particularly the diverse genome structures of newly identified picornaviruses, is a significant leap forward in understanding the crustacean virology. This knowledge is crucial for managing disease risks in aquaculture and maintaining the balance of marine ecosystems.
Keywords: crustaceans; diversity; evolution; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Diversity and connectedness of brine shrimp viruses in global hypersaline ecosystems.Sci China Life Sci. 2024 Jan;67(1):188-203. doi: 10.1007/s11427-022-2366-8. Epub 2023 Oct 30. Sci China Life Sci. 2024. PMID: 37922067
-
Viromes in marine ecosystems reveal remarkable invertebrate RNA virus diversity.Sci China Life Sci. 2022 Feb;65(2):426-437. doi: 10.1007/s11427-020-1936-2. Epub 2021 Jun 17. Sci China Life Sci. 2022. PMID: 34156600
-
Diverse RNA Viruses Associated with Diatom, Eustigmatophyte, Dinoflagellate, and Rhodophyte Microalgae Cultures.J Virol. 2022 Oct 26;96(20):e0078322. doi: 10.1128/jvi.00783-22. Epub 2022 Oct 3. J Virol. 2022. PMID: 36190242 Free PMC article.
-
A taxonomic review of viruses infecting crustaceans with an emphasis on wild hosts.J Invertebr Pathol. 2017 Jul;147:86-110. doi: 10.1016/j.jip.2017.01.010. Epub 2017 Jan 30. J Invertebr Pathol. 2017. PMID: 28153770 Review.
-
RNA Viruses in Aquatic Ecosystems through the Lens of Ecological Genomics and Transcriptomics.Viruses. 2022 Mar 28;14(4):702. doi: 10.3390/v14040702. Viruses. 2022. PMID: 35458432 Free PMC article. Review.
Cited by
-
Surveillance and Genomic Evolution of Infectious Precocity Virus (IPV) from 2011 to 2024.Viruses. 2025 Mar 15;17(3):425. doi: 10.3390/v17030425. Viruses. 2025. PMID: 40143352 Free PMC article.
-
Identification of novel hepaciviruses and Sylvilagus-associated viruses via metatranscriptomics in North American lagomorphs.Virus Evol. 2025 Jul 2;11(1):veaf050. doi: 10.1093/ve/veaf050. eCollection 2025. Virus Evol. 2025. PMID: 40687189
References
-
- FAO . 2022. The state of world fisheries and aquaculture 2022. Towards blue transformation. FAO, Rome.
-
- Stentiford GD, Neil DM, Peeler EJ, Shields JD, Small HJ, Flegel TW, Vlak JM, Jones B, Morado F, Moss S, Lotz J, Bartholomay L, Behringer DC, Hauton C, Lightner DV. 2012. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J Invertebr Pathol 110:141–157. doi:10.1016/j.jip.2012.03.013 - DOI - PubMed
-
- Britannica . 2021. The editors of encyclopaedia. “crustacean summary.” Encyclopedia Britannica
-
- Stentiford GD, Bateman IJ, Hinchliffe SJ, Bass D, Hartnell R, Santos EM, Devlin MJ, Feist SW, Taylor NGH, Verner-Jeffreys DW, van Aerle R, Peeler EJ, Higman WA, Smith L, Baines R, Behringer DC, Katsiadaki I, Froehlich HE, Tyler CR. 2020. Sustainable aquaculture through the one health iens. N Food 1:468–474. doi:10.1038/s43016-020-0127-5 - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

