Salmonella Detection in Food Using a HEK-hTLR5 Reporter Cell-Based Sensor
- PMID: 39329819
- PMCID: PMC11430776
- DOI: 10.3390/bios14090444
Salmonella Detection in Food Using a HEK-hTLR5 Reporter Cell-Based Sensor
Abstract
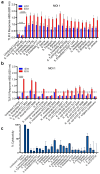
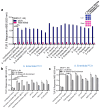
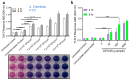
The development of a rapid, sensitive, specific method for detecting foodborne pathogens is paramount for supplying safe food to enhance public health safety. Despite the significant improvement in pathogen detection methods, key issues are still associated with rapid methods, such as distinguishing living cells from dead, the pathogenic potential or health risk of the analyte at the time of consumption, the detection limit, and the sample-to-result. Mammalian cell-based assays analyze pathogens' interaction with host cells and are responsive only to live pathogens or active toxins. In this study, a human embryonic kidney (HEK293) cell line expressing Toll-Like Receptor 5 (TLR-5) and chromogenic reporter system (HEK dual hTLR5) was used for the detection of viable Salmonella in a 96-well tissue culture plate. This cell line responds to low concentrations of TLR5 agonist flagellin. Stimulation of TLR5 ligand activates nuclear factor-kB (NF-κB)-linked alkaline phosphatase (AP-1) signaling cascade inducing the production of secreted embryonic alkaline phosphatase (SEAP). With the addition of a ρ-nitrophenyl phosphate as a substrate, a colored end product representing a positive signal is quantified. The assay's specificity was validated with the top 20 Salmonella enterica serovars and 19 non-Salmonella spp. The performance of the assay was also validated with spiked food samples. The total detection time (sample-to-result), including shortened pre-enrichment (4 h) and selective enrichment (4 h) steps with artificially inoculated outbreak-implicated food samples (chicken, peanut kernel, peanut butter, black pepper, mayonnaise, and peach), was 15 h when inoculated at 1-100 CFU/25 g sample. These results show the potential of HEK-DualTM hTLR5 cell-based functional biosensors for the rapid screening of Salmonella.
Keywords: HEK-dual hTLR 5; Salmonella; cell-based sensor; detection; flagella; food safety; immunomagnetic separation.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Bhunia A.K., editor. Foodborne Microbial Pathogens: Mechanisms and Pathogenesis. Springer; New York, NY, USA: 2018. Salmonella enterica; pp. 271–287. - DOI
-
- Teklemariam A.D., Al-Hindi R.R., Albiheyri R.S., Alharbi M.G., Alghamdi M.A., Filimban A.A., Al Mutiri A.S., Al-Alyani A.M., Alseghayer M.S., Almaneea A.M., et al. Human Salmonellosis: A Continuous Global Threat in the Farm-to-Fork Food Safety Continuum. Foods. 2023;12:1756. doi: 10.3390/foods12091756. - DOI - PMC - PubMed
-
- O’Bryan C.A., Ricke S.C., Marcy J.A. Public health impact of Salmonella spp. on raw poultry: Current concepts and future prospects in the United States. Food Control. 2022;132:108539. doi: 10.1016/j.foodcont.2021.108539. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

