Disulfidptosis: A New Target for Parkinson's Disease and Cancer
- PMID: 39329952
- PMCID: PMC11430384
- DOI: 10.3390/cimb46090600
Disulfidptosis: A New Target for Parkinson's Disease and Cancer
Abstract
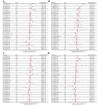
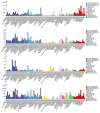
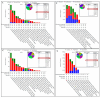
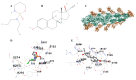
Recent studies have uncovered intriguing connections between Parkinson's disease (PD) and cancer, two seemingly distinct disease categories. Disulfidptosis has garnered attention as a novel form of regulated cell death that is implicated in various pathological conditions, including neurodegenerative disorders and cancer. Disulfidptosis involves the dysregulation of intracellular redox homeostasis, leading to the accumulation of disulfide bonds and subsequent cell demise. This has sparked our interest in exploring common molecular mechanisms and genetic factors that may be involved in the relationship between neurodegenerative diseases and tumorigenesis. The Gene4PD database was used to retrieve PD differentially expressed genes (DEGs), the biological functions of differential expression disulfidptosis-related genes (DEDRGs) were analyzed, the ROCs of DEDRGs were analyzed using the GEO database, and the expression of DEDRGs was verified by an MPTP-induced PD mouse model in vivo. Then, the DEDRGs in more than 9000 samples of more than 30 cancers were comprehensively and systematically characterized by using multi-omics analysis data. In PD, we obtained a total of four DEDRGs, including ACTB, ACTN4, INF2, and MYL6. The enriched biological functions include the regulation of the NF-κB signaling pathway, mitochondrial function, apoptosis, and tumor necrosis factor, and these genes are rich in different brain regions. In the MPTP-induced PD mouse model, the expression of ACTB was decreased, while the expression of ACTN4, INF2, and MYL6 was increased. In pan-cancer, the high expression of ACTB, ACTN4, and MYL6 in GBMLGG, LGG, MESO, and LAML had a poor prognosis, and the high expression of INF2 in LIHC, LUAD, UVM, HNSC, GBM, LAML, and KIPAN had a poor prognosis. Our study showed that these genes were more highly infiltrated in Macrophages, NK cells, Neutrophils, Eosinophils, CD8 T cells, T cells, T helper cells, B cells, dendritic cells, and mast cells in pan-cancer patients. Most substitution mutations were G-to-A transitions and C-to-T transitions. We also found that miR-4298, miR-296-3p, miR-150-3p, miR-493-5p, and miR-6742-5p play important roles in cancer and PD. Cyclophosphamide and ethinyl estradiol may be potential drugs affected by DEDRGs for future research. This study found that ACTB, ACTN4, INF2, and MYL6 are closely related to PD and pan-cancer and can be used as candidate genes for the diagnosis, prognosis, and therapeutic biomarkers of neurodegenerative diseases and cancers.
Keywords: Parkinson’s disease; disulfidptosis; immune infiltration; pan-cancer; prognosis.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures









References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

