Impacts of DROSHA (rs10719) and DICER (rs3742330) Variants on Breast Cancer Risk and Their Distribution in Blood and Tissue Samples of Egyptian Patients
- PMID: 39329954
- PMCID: PMC11430749
- DOI: 10.3390/cimb46090602
Impacts of DROSHA (rs10719) and DICER (rs3742330) Variants on Breast Cancer Risk and Their Distribution in Blood and Tissue Samples of Egyptian Patients
Abstract
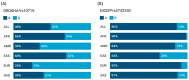
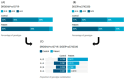
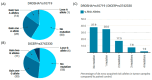
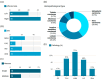
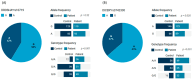
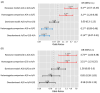
MicroRNAs (miRNAs) are small, noncoding RNAs that regulate gene expression and play critical roles in tumorigenesis. Genetic variants in miRNA processing genes, DROSHA and DICER, have been implicated in cancer susceptibility and progression in various populations. However, their role in Egyptian patients with breast cancer (BC) remains unexplored. This study aims to investigate the association of DROSHA rs10719 and DICER rs3742330 polymorphisms with BC risk and clinical outcomes. This case-control study included 209 BC patients and 106 healthy controls. Genotyping was performed using TaqMan assays in blood, tumor tissue, and adjacent non-cancerous tissue samples. Associations were analyzed using logistic regression and Fisher's exact test. The DROSHA rs10719 AA genotype was associated with a 3.2-fold increased risk (95%CI = 1.23-9.36, p < 0.001), and the DICER rs3742330 GG genotype was associated with a 3.51-fold increased risk (95%CI = 1.5-8.25, p = 0.001) of BC. Minor allele frequencies were 0.42 for rs10719 A and 0.37 for rs3742330 G alleles. The risk alleles were significantly more prevalent in tumor tissue than adjacent normal tissue (rs10719 A: 40.8% vs. 0%; rs3742330 G: 42.7% vs. 0%; p < 0.001). However, no significant associations were observed with clinicopathological features or survival outcomes over a median follow-up of 17 months. In conclusion, DROSHA rs10719 and DICER rs3742330 polymorphisms are associated with increased BC risk and more prevalent in tumor tissue among our cohort, suggesting a potential role in miRNA dysregulation during breast tumorigenesis. These findings highlight the importance of miRNA processing gene variants in BC susceptibility and warrant further validation in larger cohorts and different ethnic populations.
Keywords: DICER; DROSHA; breast cancer; microRNA; polymorphism; susceptibility.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
[The Association Between the Polymorphisms of miRNA Biogenesis Related Genes(DICER,DROSHA and RAN)and Unexplained Recurrent Spontaneous Abortion in Chinese Women].Sichuan Da Xue Xue Bao Yi Xue Ban. 2017 Nov;48(6):880-885. Sichuan Da Xue Xue Bao Yi Xue Ban. 2017. PMID: 29260525 Chinese.
-
Genetic variants in microRNA machinery genes are associated [corrected] with idiopathic recurrent pregnancy loss risk.PLoS One. 2014 Apr 25;9(4):e95803. doi: 10.1371/journal.pone.0095803. eCollection 2014. PLoS One. 2014. PMID: 24769857 Free PMC article.
-
[Association of polymorphisms of miRNA biogenesis related genes DICER and DROSHA with azoospermia].Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2016 Jun;33(3):365-8. doi: 10.3760/cma.j.issn.1003-9406.2016.03.020. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2016. PMID: 27264823 Chinese.
-
DROSHA rs10719 and DICER1 rs3742330 polymorphisms in endometriosis and different diseases: Case-control and review studies.Exp Mol Pathol. 2021 Apr;119:104616. doi: 10.1016/j.yexmp.2021.104616. Epub 2021 Jan 31. Exp Mol Pathol. 2021. PMID: 33535080 Review.
-
Association of miRNA biosynthesis genes DROSHA and DGCR8 polymorphisms with cancer susceptibility: a systematic review and meta-analysis.Biosci Rep. 2018 Jun 27;38(3):BSR20180072. doi: 10.1042/BSR20180072. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29654164 Free PMC article.
References
-
- Buocikova V., Rios-Mondragon I., Pilalis E., Chatziioannou A., Miklikova S., Mego M., Pajuste K., Rucins M., Yamani N.E., Longhin E.M., et al. Epigenetics in Breast Cancer Therapy-New Strategies and Future Nanomedicine Perspectives. Cancers. 2020;12:3622. doi: 10.3390/cancers12123622. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

