Genomic Sequencing and Functional Analysis of the Ex-Type Strain of Malbranchea zuffiana
- PMID: 39330360
- PMCID: PMC11433161
- DOI: 10.3390/jof10090600
Genomic Sequencing and Functional Analysis of the Ex-Type Strain of Malbranchea zuffiana
Abstract
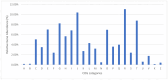
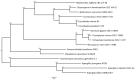
Malbranchea is a genus within the order Onygenales (phylum Ascomycota) that includes predominantly saprobic cosmopolitan species. Despite its ability to produce diverse secondary metabolites, no genomic data for Malbranchea spp. are currently available in databases. Therefore, in this study, we obtained, assembled, and annotated the genomic sequence of the ex-type strain of Malbranchea zuffiana (CBS 219.58). For the genomic sequencing, we employed both the Illumina and PacBio platforms, followed by hybrid assembly using MaSuRCA. Quality assessment of the assembly was performed using QUAST and BUSCO tools. Annotation was conducted using BRAKER2, and functional annotation was completed with InterProScan. The resulting genome was of high quality, with a size of 26.46 Mbp distributed across 38 contigs and a BUSCO completion rate of 95.7%, indicating excellent contiguity and assembly completeness. A total of 8248 protein-encoding genes were predicted, with functional annotations assigned to 73.9% of them. Moreover, 82 genes displayed homology with entries in the Pathogen Host Interactions (PHI) database, while 494 genes exhibited similarity to entries in the Carbohydrate-Active Enzymes (CAZymes) database. Furthermore, 30 biosynthetic gene clusters (BGCs) were identified, suggesting significant potential for the biosynthesis of diverse secondary metabolites. Comparative functional analysis with closely related species unveiled a considerable abundance of domains linked to enzymes involved in keratin degradation, alongside a restricted number of domains associated with enzymes engaged in plant cell wall degradation in all studied species of the Onygenales. This genome-based elucidation not only enhances our comprehension of the biological characteristics of M. zuffiana but also furnishes valuable insights for subsequent investigations concerning Malbranchea species and the order Onygenales.
Keywords: Malbranchea zuffiana; Onygenales; genes; genome assembly; keratin degradation; phylogenomics; proteins.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Hybrid De Novo Whole-Genome Assembly, Annotation, and Identification of Secondary Metabolite Gene Clusters in the Ex-Type Strain of Chrysosporium keratinophilum.J Fungi (Basel). 2023 Mar 23;9(4):389. doi: 10.3390/jof9040389. J Fungi (Basel). 2023. PMID: 37108844 Free PMC article.
-
Genome description of Phlebia radiata 79 with comparative genomics analysis on lignocellulose decomposition machinery of phlebioid fungi.BMC Genomics. 2019 May 28;20(1):430. doi: 10.1186/s12864-019-5817-8. BMC Genomics. 2019. PMID: 31138126 Free PMC article.
-
Sequencing and Functional Annotation of the Whole Genome of Shiraia bambusicola.G3 (Bethesda). 2020 Jan 7;10(1):23-35. doi: 10.1534/g3.119.400694. G3 (Bethesda). 2020. PMID: 31712259 Free PMC article.
-
Sequencing and functional annotation of the whole genome of the filamentous fungus Aspergillus westerdijkiae.BMC Genomics. 2016 Aug 15;17(1):633. doi: 10.1186/s12864-016-2974-x. BMC Genomics. 2016. PMID: 27527502 Free PMC article.
-
Whole genome sequencing and annotation of Scleroderma yunnanense, the only edible Scleroderma species.Genomics. 2023 Nov;115(6):110727. doi: 10.1016/j.ygeno.2023.110727. Epub 2023 Oct 14. Genomics. 2023. PMID: 37839651 Review.
Cited by
-
Metabolic Blockade-Based Genome Mining of Malbranchea circinata SDU050: Discovery of Diverse Secondary Metabolites.Mar Drugs. 2025 Jan 20;23(1):50. doi: 10.3390/md23010050. Mar Drugs. 2025. PMID: 39852552 Free PMC article.
References
-
- Saccardo P. Fungi Gallici lecti a Cl. viris P. Brunaud, C.C. Gillet, Abb. Letendre, A. Malbranche, J. Therry & Dom. Libert. Ser. IV. Michelia. 1882;2:583–648.
-
- Cooney D.G., Emerson R. In: Thermophilic Fungi: An Account of Their Biology. Freeman W.H., editor. Activities and Classification; San Francisco, CA, USA: London, UK: 1964.
-
- Sigler L., Carmichael J. Taxonomy of Malbranchea and some other hyphomycetes with arthroconidia. Mycotaxon. 1976;4:349–488.
-
- Sigler L., Hambleton S., Flis A.L., Paré J.A. Auxarthron teleomorphs for Malbranchea filamentosa and Malbranchea albolutea and relationships within Auxarthron. Stud. Mycol. 2002;47:111–122.
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

