Comparative Genomic Analyses of Colletotrichum lindemuthianum Pathotypes with Different Virulence Levels and Lifestyles
- PMID: 39330411
- PMCID: PMC11432805
- DOI: 10.3390/jof10090651
Comparative Genomic Analyses of Colletotrichum lindemuthianum Pathotypes with Different Virulence Levels and Lifestyles
Abstract
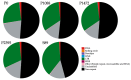
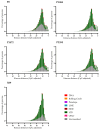
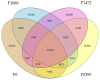
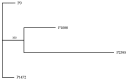
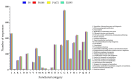
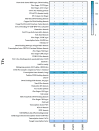
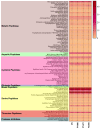
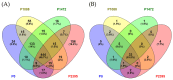
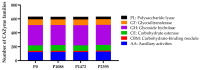
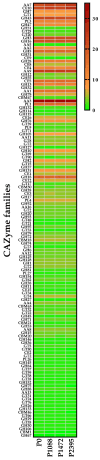
Colletotrichum lindemuthianum is the most frequent pathogenic fungus of the common bean Phaseolus vulgaris. This filamentous fungus employs a hemibiotrophic nutrition/infection strategy, which is characteristic of many Colletotrichum species. Due to host-pathogen coevolution, C. lindemuthianum includes pathotypes with a diversity of virulence against differential common bean varieties. In this study, we performed comparative genomic analyses on three pathotypes with different virulence levels and a non-pathogenic pathotype, isolated from different geographical areas in Mexico. Our results revealed large genomes with high transposable element contents that have undergone expansions, generating intraspecific diversity. All the pathotypes exhibited a similar number of clusters of orthologous genes (COGs) and Gene Ontology (GO) terms. TFomes contain families that are typical in fungal genomes; however, they show different contents between pathotypes, mainly in transcription factors with the fungal-specific TF and Zn2Cys6 domains. Peptidase families mainly contain abundant serine peptidases, metallopeptidases, and cysteine peptidases. In the secretomes, the number of genes differed between the pathotypes, with a high percentage of candidate effectors. Both the virulence gene and CAZyme gene content for each pathotype was abundant and diverse, and the latter was enriched in hemicellulolytic enzymes. We provide new insights into the nature of intraspecific diversity among C. lindemuthianum pathotypes and the origin of their ability to rapidly adapt to genetic changes in its host and environmental conditions.
Keywords: CAZymes; TFomes; effectors; genomic; intraspecific diversity; pathotypes; proteases; transposable elements; virulence genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Differential Carbon Catabolite Repression and Hemicellulolytic Ability among Pathotypes of Colletotrichum lindemuthianum against Natural Plant Substrates.J Fungi (Basel). 2024 Jun 5;10(6):406. doi: 10.3390/jof10060406. J Fungi (Basel). 2024. PMID: 38921392 Free PMC article.
-
Analysis of Pathotypes of Colletotrichum lindemuthianum Found in the Central Region of Mexico and Resistance in Elite Germ Plasm of Phaseolus vulgaris.Plant Dis. 2004 Feb;88(2):152-156. doi: 10.1094/PDIS.2004.88.2.152. Plant Dis. 2004. PMID: 30812421
-
What lies behind the large genome of Colletotrichum lindemuthianum.Front Fungal Biol. 2024 Oct 15;5:1459229. doi: 10.3389/ffunb.2024.1459229. eCollection 2024. Front Fungal Biol. 2024. PMID: 39473581 Free PMC article.
-
Phaseolus vulgaris-Colletotrichum lindemuthianum Pathosystem in the Post-Genomic Era: An Update.Curr Microbiol. 2022 Jan 4;79(2):36. doi: 10.1007/s00284-021-02711-6. Curr Microbiol. 2022. PMID: 34982236 Review.
-
Updates on defining and detecting diarrheagenic Escherichia coli pathotypes.Curr Opin Infect Dis. 2020 Oct;33(5):372-380. doi: 10.1097/QCO.0000000000000665. Curr Opin Infect Dis. 2020. PMID: 32773499 Free PMC article. Review.
Cited by
-
Uncovering the Host Range-Lifestyle Relationship in the Endophytic and Anthracnose Pathogenic Genus Colletotrichum.Microorganisms. 2025 Feb 16;13(2):428. doi: 10.3390/microorganisms13020428. Microorganisms. 2025. PMID: 40005793 Free PMC article. Review.
References
-
- Rava C., Purchio A., Sartorato A. Caracterização de patótipos de Colletotrichum lindemuthianum que ocorrem em algumas regiões produtoras de feijoeiro comum. Fitopatol. Bras. 1994;19:167–172.
-
- Padder B., Sharma P., Sharma P., Sharma O., Kapoor V. Genetic diversity and gene flow estimates among five populations of Colletotrichum lindemuthianum across Himachal Pradesh. Physiol. Mol. Plant Path. 2007;70:8–12. doi: 10.1016/j.pmpp.2007.05.003. - DOI
-
- Sharma P., Sharma O., Padder B., Kapil R. Yield loss assessment in common bean due to anthracnose (Colletotrichum lindemuthianum) under sub temperate conditions of North-Western Himalayas. Indian Phytopath. 2008;61:323.
-
- Singh S.P., Schwartz H. Breeding common bean for resistance to diseases: A review. Crop Sci. 2010;50:2199–2223. doi: 10.2135/cropsci2009.03.0163. - DOI
Grants and funding
- SEP-CONAHCYT-CB2017-2018 A1 S-27298/Consejo Nacional de Humanidades, Ciencias y Tecnologías, México
- Scholarship No. 792799/Consejo Nacional de Humanidades, Ciencias y Tecnologías, México
- Scholarship No. 792802/Consejo Nacional de Humanidades, Ciencias y Tecnologías, México
- 2020-2023/Coordinación de la Investigación Científica, Universidad Michoacana de San Nicolás de Hidalgo
LinkOut - more resources
Full Text Sources
Miscellaneous

