Utility of an Untargeted Metabolomics Approach Using a 2D GC-GC-MS Platform to Distinguish Relapsing and Progressive Multiple Sclerosis
- PMID: 39330500
- PMCID: PMC11434588
- DOI: 10.3390/metabo14090493
Utility of an Untargeted Metabolomics Approach Using a 2D GC-GC-MS Platform to Distinguish Relapsing and Progressive Multiple Sclerosis
Abstract
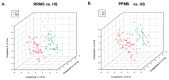
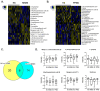
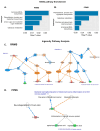
Multiple sclerosis (MS) is the most common inflammatory neurodegenerative disease of the central nervous system (CNS) in young adults and results in progressive neurological defects. The relapsing-remitting phenotype (RRMS) is the most common disease course in MS, which ultimately progresses to secondary progressive MS (SPMS), while primary progressive MS (PPMS) is a type of MS that worsens gradually over time without remissions. There is a gap in knowledge regarding whether the relapsing form can be distinguished from the progressive course, or healthy subjects (HS) based on an altered serum metabolite profile. In this study, we performed global untargeted metabolomics with the 2D GC-GC-MS platform to identify altered metabolites between RRMS, PPMS, and HS. We profiled 235 metabolites in the serum of patients with RRMS (n = 41), PPMS (n = 31), and HS (n = 91). A comparison of RRMS and HS patients revealed 22 significantly altered metabolites at p < 0.05 (false-discovery rate [FDR] = 0.3). The PPMS and HS comparisons revealed 28 altered metabolites at p < 0.05 (FDR = 0.2). Pathway analysis using MetaboAnalyst revealed enrichment of four metabolic pathways in both RRMS and PPMS (hypergeometric test p < 0.05): (1) galactose metabolism; (2) amino sugar and nucleotide sugar metabolism; (3) phenylalanine, tyrosine, and tryptophan biosynthesis; and (4) aminoacyl-tRNA biosynthesis. The Qiagen IPA enrichment test identified the sulfatase 2 (SULF2) (p = 0.0033) and integrin subunit beta 1 binding protein 1 (ITGB1BP1) (p = 0.0067) genes as upstream regulators of altered metabolites in the RRMS vs. HS groups. However, in the PPMS vs. HS comparison, valine was enriched in the neurodegeneration of brain cells (p = 0.05), and heptadecanoic acid, alpha-ketoisocaproic acid, and glycerol participated in inflammation in the CNS (p = 0.03). Overall, our study suggests that RRMS and PPMS may contribute metabolic fingerprints in the form of unique altered metabolites for discriminating MS disease from HS, with the potential for constructing a metabolite panel for progressive autoimmune diseases such as MS.
Keywords: GC-GC-MS; PPMS; RRMS; metabolomics; multiple sclerosis.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures


Update of
-
Utility of an untargeted metabolomics approach using a 2D GC-GC-MS platform to distinguish relapsing and progressive multiple sclerosis.bioRxiv [Preprint]. 2024 Feb 10:2024.02.07.579252. doi: 10.1101/2024.02.07.579252. bioRxiv. 2024. Update in: Metabolites. 2024 Sep 11;14(9):493. doi: 10.3390/metabo14090493. PMID: 38370675 Free PMC article. Updated. Preprint.
Similar articles
-
Utility of an untargeted metabolomics approach using a 2D GC-GC-MS platform to distinguish relapsing and progressive multiple sclerosis.bioRxiv [Preprint]. 2024 Feb 10:2024.02.07.579252. doi: 10.1101/2024.02.07.579252. bioRxiv. 2024. Update in: Metabolites. 2024 Sep 11;14(9):493. doi: 10.3390/metabo14090493. PMID: 38370675 Free PMC article. Updated. Preprint.
-
Multiple sclerosis by phenotype in Germany.Mult Scler Relat Disord. 2022 Jan;57:103326. doi: 10.1016/j.msard.2021.103326. Epub 2021 Oct 10. Mult Scler Relat Disord. 2022. PMID: 35158442
-
Serious infections in patients with relapsing and progressive forms of multiple sclerosis: A German claims data study.Mult Scler Relat Disord. 2022 Dec;68:104245. doi: 10.1016/j.msard.2022.104245. Epub 2022 Oct 17. Mult Scler Relat Disord. 2022. PMID: 36306609
-
Gray Matter Atrophy in the Cortico-Striatal-Thalamic Network and Sensorimotor Network in Relapsing-Remitting and Primary Progressive Multiple Sclerosis.Neuropsychol Rev. 2021 Dec;31(4):703-720. doi: 10.1007/s11065-021-09479-3. Epub 2021 Feb 13. Neuropsychol Rev. 2021. PMID: 33582965 Review.
-
Recent Advances in Metabolomics and Lipidomics Studies in Human and Animal Models of Multiple Sclerosis.Metabolites. 2024 Oct 13;14(10):545. doi: 10.3390/metabo14100545. Metabolites. 2024. PMID: 39452926 Free PMC article. Review.
Cited by
-
Artificial neural network-based prediction of multiple sclerosis using blood-based metabolomics data.Mult Scler Relat Disord. 2024 Dec;92:105942. doi: 10.1016/j.msard.2024.105942. Epub 2024 Oct 15. Mult Scler Relat Disord. 2024. PMID: 39471746
-
Metabolic Characterization of Cerebrospinal Fluid for Patients With Autoimmune Encephalitis: A Preliminary Study.CNS Neurosci Ther. 2025 Jan;31(1):e70203. doi: 10.1111/cns.70203. CNS Neurosci Ther. 2025. PMID: 39749658 Free PMC article.
-
Immune-responsive gene-1: The mitochondrial Key to Th17 Cell Pathogenicity in CNS Autoimmunity.bioRxiv [Preprint]. 2025 Feb 10:2023.12.24.573264. doi: 10.1101/2023.12.24.573264. bioRxiv. 2025. Update in: Proc Natl Acad Sci U S A. 2025 Aug 12;122(32):e2427052122. doi: 10.1073/pnas.2427052122. PMID: 38234838 Free PMC article. Updated. Preprint.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

