Implementation of Machine Learning-Based System for Early Diagnosis of Feline Mammary Carcinomas through Blood Metabolite Profiling
- PMID: 39330508
- PMCID: PMC11433869
- DOI: 10.3390/metabo14090501
Implementation of Machine Learning-Based System for Early Diagnosis of Feline Mammary Carcinomas through Blood Metabolite Profiling
Abstract
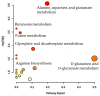
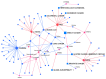
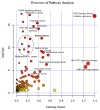
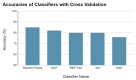
Background: Feline mammary carcinoma (FMC) is a prevalent and fatal carcinoma that predominantly affects unspayed female cats. FMC is the third most common carcinoma in cats but is still underrepresented in research. Current diagnosis methods include physical examinations, imaging tests, and fine-needle aspiration. The diagnosis through these methods is sometimes delayed and unreliable, leading to increased chances of mortality. Objectives: The objective of this study was to identify the biomarkers, including blood metabolites and genes, related to feline mammary carcinoma, study their relationships, and develop a machine learning (ML) model for the early diagnosis of the disease. Methods: We analyzed the blood metabolites of felines with mammary carcinoma using the pathway analysis feature in MetaboAnalyst software, v. 5.0. We utilized machine-learning (ML) methods to recognize FMC using the blood metabolites of sick patients. Results: The metabolic pathways that were elucidated to be associated with this disease include alanine, aspartate and glutamate metabolism, Glutamine and glutamate metabolism, Arginine biosynthesis, and Glycerophospholipid metabolism. Furthermore, we also elucidated several genes that play a significant role in the development of FMC, such as ERBB2, PDGFA, EGFR, FLT4, ERBB3, FIGF, PDGFC, PDGFB through STRINGdb, a database of known and predicted protein-protein interactions, and MetaboAnalyst 5.0. The best-performing ML model was able to predict metabolite class with an accuracy of 85.11%. Conclusion: Our findings demonstrate that the identification of the biomarkers associated with FMC and the affected metabolic pathways can aid in the early diagnosis of feline mammary carcinoma.
Keywords: early diagnosis; feline mammary cancer; machine learning; metabolomics.
Conflict of interest statement
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article. Igor Tsigelny and Valentina Kouznetsova are the employees of BiAna. The paper reflects the views of the scientists, and not the company.
Figures



Similar articles
-
Recognition of early and late stages of bladder cancer using metabolites and machine learning.Metabolomics. 2019 Jun 20;15(7):94. doi: 10.1007/s11306-019-1555-9. Metabolomics. 2019. PMID: 31222577
-
Feline Mammary Cancer.Vet Pathol. 2017 Jan;54(1):32-43. doi: 10.1177/0300985816650243. Epub 2016 Jul 11. Vet Pathol. 2017. PMID: 27281014 Free PMC article.
-
Serum SDF-1 levels are a reliable diagnostic marker of feline mammary carcinoma, discriminating HER2-overexpressing tumors from other subtypes.Oncotarget. 2017 Nov 11;8(62):105775-105789. doi: 10.18632/oncotarget.22398. eCollection 2017 Dec 1. Oncotarget. 2017. PMID: 29285291 Free PMC article.
-
Emerging Biomarkers and Targeted Therapies in Feline Mammary Carcinoma.Vet Sci. 2021 Aug 11;8(8):164. doi: 10.3390/vetsci8080164. Vet Sci. 2021. PMID: 34437486 Free PMC article. Review.
-
Adipocyte-Derived Adipokines and Other Obesity-Associated Molecules in Feline Mammary Cancer.Biomedicines. 2023 Aug 19;11(8):2309. doi: 10.3390/biomedicines11082309. Biomedicines. 2023. PMID: 37626804 Free PMC article. Review.
Cited by
-
Prognostic Insights in Feline Mammary Carcinomas: Clinicopathological Factors and the Proposal of a New Staging System.Animals (Basel). 2025 Mar 10;15(6):779. doi: 10.3390/ani15060779. Animals (Basel). 2025. PMID: 40150308 Free PMC article.
References
-
- Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. doi: 10.1021/ci00057a005. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

