Investigating the Mechanisms of 15-PGDH Inhibitor SW033291 in Improving Type 2 Diabetes Mellitus: Insights from Metabolomics and Transcriptomics
- PMID: 39330516
- PMCID: PMC11434390
- DOI: 10.3390/metabo14090509
Investigating the Mechanisms of 15-PGDH Inhibitor SW033291 in Improving Type 2 Diabetes Mellitus: Insights from Metabolomics and Transcriptomics
Abstract
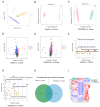
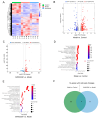
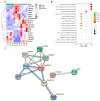
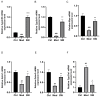
This study focused on exploring the effects of SW033291, an inhibitor of 15-hydroxyprostaglandin dehydrogenase, on type 2 diabetes mellitus (T2DM) mice from a comprehensive perspective. Studies have demonstrated that SW033291 benefits tissue repair, organ function, and muscle mass in elderly mice. Our recent investigation initially reported the beneficial effect of SW033291 on T2DM progression. Herein, we used a T2DM mouse model induced by a high-fat diet and streptozotocin injection. Then, serum and liver metabolomics, as well as liver transcriptomic analyses, were performed to provide a systematic perspective of the SW033291-ameliorated T2DM. The results indicate SW033291 improved T2DM by regulating steroid hormone biosynthesis and linoleic/arachidonic acid metabolism. Furthermore, integrated transcriptomic and metabolomic analyses suggested that key genes and metabolites such as Cyp2c55, Cyp3a11, Cyp21a1, Myc, Gstm1, Gstm3, 9,10-dihydroxyoctadecenoic acid, 11-dehydrocorticosterone, and 12,13-dihydroxy-9Z-octadecenoic acid played crucial roles in these pathways. qPCR analysis validated the significant decreases in the hepatic gene expressions of Cyp2c55, Cyp3a11, Myc, Gstm1, and Gstm3 in the T2DM mice, which were reversed following SW033291 treatment. Meanwhile, the elevated mRNA level of Cyp21a1 in T2DM mice was decreased after SW033291 administration. Taken together, our findings suggest that SW033291 has promising potential in alleviating T2DM and could be a novel therapeutic candidate.
Keywords: 15-hydroxyprostaglandin dehydrogenase; SW033291; metabolomics; transcriptomics; type 2 diabetes mellitus.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures


References
-
- Sun H., Saeedi P., Karuranga S., Pinkepank M., Ogurtsova K., Duncan B.B., Stein C., Basit A., Chan J.C.N., Mbanya J.C., et al. IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res. Clin. Pract. 2022;183:109119. doi: 10.1016/j.diabres.2021.109119. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

