First whole genome sequencing and analysis of human parechovirus type 3 causing a healthcare-associated outbreak among neonates in Hungary
- PMID: 39331310
- PMCID: PMC11608160
- DOI: 10.1007/s10096-024-04950-4
First whole genome sequencing and analysis of human parechovirus type 3 causing a healthcare-associated outbreak among neonates in Hungary
Abstract
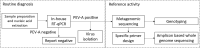
Purpose: In November 2023, the National Reference Laboratory for Enteroviruses (Budapest, Hungary) received stool, pharyngeal swab and cerebrospinal fluid samples from five newborns suspected of having human parechovirus (PEV-A) infection. The neonates were born in the same hospital and presented with fever and sepsis-like symptoms at 8-9 days of age, and three of them showed symptoms consistent with central nervous system involvement. PEV-A positivity was confirmed by quantitative reverse transcription polymerase chain reaction.
Methods: To determine the PEV-A genotype responsible for the infections, fecal samples of four neonates were subjected to metagenomic sequencing. For further analyses, amplicon-based whole genome sequencing was performed directly from the clinical samples.
Results: On the basis of whole genome analysis, sequences were allocated to PEV-A genotype 3 (PEV-A3) and consensus sequences were identical. Two ambiguities were identified in the viral protein 1 (VP1) region of all sequences at a frequency of 17.7-53.7%, indicating the simultaneous presence of at least two quasispecies in the clinical samples. The phylogenetic analysis and similarity plotting showed that all sequences clustered without any topological inconsistencies between the P1 capsid and P2, P3 non-capsid regions, suggesting that recombination events during evolution were unlikely.
Conclusion: Our findings suggest that the apparent cluster of cases were microbiologically related, and the results may also inform future investigations on the evolution and pathogenicity of PEV-A3 infections.
Keywords: Genotyping; Human parechovirus 3; Infection of neonates; Phylogenetic analysis; Whole genome sequencing.
© 2024. The Author(s).
Conflict of interest statement
Declarations. Ethical approval: Ethics Committee approval was not required as the Hungarian legislation on handling of personal health information (Law no. 1997. XLVII.) empowers the National Center for Public Health and Pharmacy to analyse data and take necessary measures in the interest of public health. Personal data have been handled in accordance with legal regulations and the Center’s data protection rules. Competing interests: The authors declare no competing interests.
Figures


Similar articles
-
Intrafamilial Transmission of Parechovirus A and Enteroviruses in Neonates and Young Infants.J Pediatric Infect Dis Soc. 2019 Dec 27;8(6):501-506. doi: 10.1093/jpids/piy079. J Pediatric Infect Dis Soc. 2019. PMID: 30184210
-
Genome Announcement of Four Parechovirus A3 Isolates from the United States of America.Int J Mol Sci. 2025 May 5;26(9):4378. doi: 10.3390/ijms26094378. Int J Mol Sci. 2025. PMID: 40362615 Free PMC article.
-
[The clinical significance of severe human parechovirus infections in newborns and infants in Hungary].Orv Hetil. 2019 Mar;160(10):386-395. doi: 10.1556/650.2019.31304. Orv Hetil. 2019. PMID: 30829060 Hungarian.
-
Diagnosis, management, and outcomes of parechovirus infections in infants: an overview.J Clin Microbiol. 2024 Jun 12;62(6):e0113923. doi: 10.1128/jcm.01139-23. Epub 2024 Apr 22. J Clin Microbiol. 2024. PMID: 38647282 Free PMC article. Review.
-
[Recent progress in the research of human parechovirus 3].Zhonghua Er Ke Za Zhi. 2013 Feb;51(2):111-4. Zhonghua Er Ke Za Zhi. 2013. PMID: 23527974 Review. Chinese. No abstract available.
Cited by
-
Evaluating enterovirus diversity among symptomatic patients in Hungary during and after easing the COVID-19 lockdown.Virol J. 2025 Jun 24;22(1):204. doi: 10.1186/s12985-025-02835-2. Virol J. 2025. PMID: 40555984 Free PMC article.
References
-
- Picornaviridae Study Group (2024) https://www.picornastudygroup.com/types/parechovirus/parechovirus_a.htm (last online access: 23
-
- Ghazi F, Hughes PJ, Hyypiä T, Stanway G (1998 Nov) Molecular analysis of human parechovirus type 2 (formerly echovirus 23). J Gen Virol 79(Pt 11):2641–2650. 10.1099/0022-1317-79-11-2641 - PubMed
-
- Pietsch C, Liebert UG (2019 Mar) Genetic diversity of human parechoviruses in stool samples, Germany. Infect Genet Evol 68:280–285. 10.1016/j.meegid.2019.01.007 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

