Poly ADP-ribose signaling is dysregulated in Huntington disease
- PMID: 39331414
- PMCID: PMC11459172
- DOI: 10.1073/pnas.2318098121
Poly ADP-ribose signaling is dysregulated in Huntington disease
Abstract
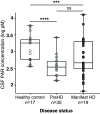
Huntington disease (HD) is a genetic neurodegenerative disease caused by cytosine, adenine, guanine (CAG) expansion in the Huntingtin (HTT) gene, translating to an expanded polyglutamine tract in the HTT protein. Age at disease onset correlates to CAG repeat length but varies by decades between individuals with identical repeat lengths. Genome-wide association studies link HD modification to DNA repair and mitochondrial health pathways. Clinical studies show elevated DNA damage in HD, even at the premanifest stage. A major DNA repair node influencing neurodegenerative disease is the PARP pathway. Accumulation of poly adenosine diphosphate (ADP)-ribose (PAR) has been implicated in Alzheimer and Parkinson diseases, as well as cerebellar ataxia. We report that HD mutation carriers have lower cerebrospinal fluid PAR levels than healthy controls, starting at the premanifest stage. Human HD induced pluripotent stem cell-derived neurons and patient-derived fibroblasts have diminished PAR response in the context of elevated DNA damage. We have defined a PAR-binding motif in HTT, detected HTT complexed with PARylated proteins in human cells during stress, and localized HTT to mitotic chromosomes upon inhibition of PAR degradation. Direct HTT PAR binding was measured by fluorescence polarization and visualized by atomic force microscopy at the single molecule level. While wild-type and mutant HTT did not differ in their PAR binding ability, purified wild-type HTT protein increased in vitro PARP1 activity while mutant HTT did not. These results provide insight into an early molecular mechanism of HD, suggesting possible targets for the design of early preventive therapies.
Keywords: Huntington’s disease; PARP1; PARylation; huntingtin; poly ADP-ribose.
Conflict of interest statement
Competing interests statement:E.J.W. reports consultancy/ advisory board memberships with Annexon, Remix Therapeutics, Hoffman La Roche Ltd., Ionis Pharmaceuticals, PTC Therapeutics, Takeda, Teitur Trophics, Triplet Therapeutics and Vico Therapeutics. All honoraria for these consultancies were paid through the offices of UCL Consultants Ltd., a wholly owned subsidiary of University College London. F.B.R. is a Medpace UK Ltd. employee and was a University College London employee during the conduct of this study. F.B.R. has provided consultancy services to GLG and F. Hoffmann-La Roche Ltd. L.M.B. currently holds consultancy contracts with Annexon Biosciences, Remix Therapeutics, and LoQus23 Therapeutics Ltd. via UCL Consultants Ltd. R.T. has past consultancy with Novartis AG, PTC Therapeutics and Mitokinin LLC.
Figures





References
-
- Djousse L., et al. , Interaction of normal and expanded CAG repeat sizes influences age at onset of Huntington disease. Am. J. Med. Genet. A 119, 279–282 (2003). - PubMed
MeSH terms
Substances
Grants and funding
- PJT-180258/Canadian Government | Canadian Institutes of Health Research (CIHR)
- T32 CA009110/CA/NCI NIH HHS/United States
- T32 GM149382/GM/NIGMS NIH HHS/United States
- R35 NS116872/NS/NINDS NIH HHS/United States
- R37 NS067525/NS/NINDS NIH HHS/United States
- PJT-168966/Canadian Government | Canadian Institutes of Health Research (CIHR)
- T32GM080189/HHS | NIH (NIH)
- MR/M008592/1/MRC_/Medical Research Council/United Kingdom
- T32 GM080189/GM/NIGMS NIH HHS/United States
- R01 GM104135/GM/NIGMS NIH HHS/United States
- MR/M008592/1/UKRI | MRC | Medical Research Council Centre for Neurodevelopmental Disorders (CNDD)
- T32-CA009110/HHS | NIH (NIH)
- MR/W026686/1/MRC_/Medical Research Council/United Kingdom
- CHDI002/CHDI Foundation (CHDI)
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

