Local volume concentration, packing domains, and scaling properties of chromatin
- PMID: 39331520
- PMCID: PMC11434620
- DOI: 10.7554/eLife.97604
Local volume concentration, packing domains, and scaling properties of chromatin
Abstract
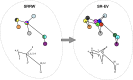
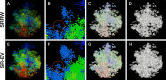
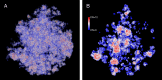
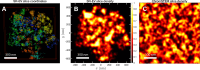
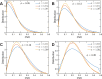
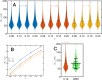
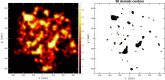
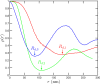
We propose the Self Returning Excluded Volume (SR-EV) model for the structure of chromatin based on stochastic rules and physical interactions. The SR-EV rules of return generate conformationally defined domains observed by single-cell imaging techniques. From nucleosome to chromosome scales, the model captures the overall chromatin organization as a corrugated system, with dense and dilute regions alternating in a manner that resembles the mixing of two disordered bi-continuous phases. This particular organizational topology is a consequence of the multiplicity of interactions and processes occurring in the nuclei, and mimicked by the proposed return rules. Single configuration properties and ensemble averages show a robust agreement between theoretical and experimental results including chromatin volume concentration, contact probability, packing domain identification and size characterization, and packing scaling behavior. Model and experimental results suggest that there is an inherent chromatin organization regardless of the cell character and resistant to an external forcing such as RAD21 degradation.
Keywords: SR-EV; chromatin; none; physics of living systems; theory.
© 2024, Carignano, Kroeger, Almassalha et al.
Conflict of interest statement
MC, MK, LA, VA, WL, EP, RN, VB, IS No competing interests declared
Figures

Update of
-
Local Volume Concentration, Packing Domains and Scaling Properties of Chromatin.Res Sq [Preprint]. 2023 Oct 17:rs.3.rs-3399177. doi: 10.21203/rs.3.rs-3399177/v1. Res Sq. 2023. Update in: Elife. 2024 Sep 27;13:RP97604. doi: 10.7554/eLife.97604. PMID: 37886531 Free PMC article. Updated. Preprint.
-
Local Volume Concentration, Packing Domains and Scaling Properties of Chromatin.ArXiv [Preprint]. 2024 Jun 13:arXiv:2310.02257v3. ArXiv. 2024. Update in: Elife. 2024 Sep 27;13:RP97604. doi: 10.7554/eLife.97604. PMID: 38495560 Free PMC article. Updated. Preprint.
References
-
- Almassalha LM, Bauer GM, Chandler JE, Gladstein S, Cherkezyan L, Stypula-Cyrus Y, Weinberg S, Zhang D, Thusgaard Ruhoff P, Roy HK, Subramanian H, Chandel NS, Szleifer I, Backman V. Label-free imaging of the native, living cellular nanoarchitecture using partial-wave spectroscopic microscopy. PNAS. 2016;113:E6372–E6381. doi: 10.1073/pnas.1608198113. - DOI - PMC - PubMed
-
- Annunziato A. DNA packaging: Nucleosomes and chromatin. Nature Education. 2008;1:26
MeSH terms
Substances
Grants and funding
- T32AI083216/National Institute of Allergy and Infectious Diseases
- U54 CA261694/CA/NCI NIH HHS/United States
- U54CA261694/CA/NCI NIH HHS/United States
- R01 CA224911/CA/NCI NIH HHS/United States
- R01CA228272/CA/NCI NIH HHS/United States
- EFMA-1830961/National Science Foundation
- R01CA225002/CA/NCI NIH HHS/United States
- T32 AI083216/AI/NIAID NIH HHS/United States
- U54 CA268084/CA/NCI NIH HHS/United States
- T32GM142604/GM/NIGMS NIH HHS/United States
- R01 CA228272/CA/NCI NIH HHS/United States
- R01CA224911/CA/NCI NIH HHS/United States
- R01 CA225002/CA/NCI NIH HHS/United States
- T32 GM142604/GM/NIGMS NIH HHS/United States
- U54CA268084/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials

