Characterization of gut symbionts from wild-caught Drosophila and other Diptera: description of Utexia brackfieldae gen. nov., sp. nov., Orbus sturtevantii sp. nov., Orbus wheelerorum sp. nov, and Orbus mooreae sp. nov
- PMID: 39331838
- PMCID: PMC11434166
- DOI: 10.1099/ijsem.0.006516
Characterization of gut symbionts from wild-caught Drosophila and other Diptera: description of Utexia brackfieldae gen. nov., sp. nov., Orbus sturtevantii sp. nov., Orbus wheelerorum sp. nov, and Orbus mooreae sp. nov
Abstract
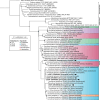
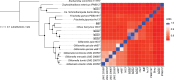
Non-culture based surveys show that the bacterial family Orbaceae is widespread in guts of insects, including wild Drosophila. Relatively few isolates have been described, and none has been described from Drosophila. We present the isolation and characterization of five strains of Orbaceae from wild-caught flies of the genera Drosophila (Diptera: Drosophilidae) and Neogriphoneura (Diptera: Lauxaniidae). Cells are generally rod-shaped, mesophilic, and measure 0.8-2.0 µm long by 0.3-0.5 µm wide. Optimal growth was observed under ambient atmosphere. Reconstruction of phylogenies from the 16S rRNA gene and from single-copy orthologs verify placement of these strains within Orbaceae. Cells exhibited similar fatty acid profiles to those of other Orbaceae. Strain lpD01T shared 74% average nucleotide identity (ANI) with its closest relatives Ca. Schmidhempelia bombi Bimp and Zophobihabitans entericus IPMB12T. Results from multiple genome-wide similarity comparisons indicate lpD01T should be classified as a novel species within a novel genus. The major respiratory quinone for lpD01T is ubiquinone Q-8. lpD02T, lpD03, lpD04T, and BiBT are more closely related to Orbus hercynius CN3T (76, 77, 76, and 77% ANI, respectively) than to other described Orbaceae. Genomic and phylogenetic analyses suggest that lpD03 and lpD04T belong to the same species and that lpD02T, lpD03/lpD04T, and BiBT are each novel species of the genus Orbus. The proposed names of these strains are Utexia brackfieldae gen. nov., sp. nov. (type strain lpD01T =NCIMB 15517T =ATCC TSD-399T), Orbus sturtevantii sp. nov (type strain lpD02T =NCIMB 15518T =ATCC TSD-400T), Orbus wheelerorum sp. nov. (type strain lpD04T =NCIMB 15520T =ATCC TSD-401T), and Orbus mooreae sp. nov (type strain BiBT=NCIMB 15516T =ATCC TSD-402T). The isolation and characterization of these strains expands the repertoire of culturable bacteria naturally associated with insects, including the model organism D. melanogaster.
Keywords: insect microbiome; Drosophila; Orbaceae; host specificity; host-associated microbiota; symbiosis.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

