Monitoring of Pseudomonas aeruginosa mutational resistome dynamics using an enrichment panel for direct sequencing of clinical samples
- PMID: 39332391
- PMCID: PMC11467565
- DOI: 10.1016/j.ebiom.2024.105367
Monitoring of Pseudomonas aeruginosa mutational resistome dynamics using an enrichment panel for direct sequencing of clinical samples
Abstract
Background: Pseudomonas aeruginosa is a major cause of hospital-acquired and chronic infections, characterised by an extraordinary capacity to develop antimicrobial resistance through the selection of chromosomal mutations, leading to treatment failure. Here, we designed and tested a hybridisation-based capture system for the enrichment of genes of interest before sequencing to monitor resistant populations genomics directly from clinical samples.
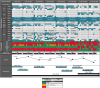
Methods: A panel for enrichment before sequencing of close to 200 genes related to P. aeruginosa antimicrobial resistance, multilocus sequence typing, mutability or virulence was designed, synthesised (KAPA HyperCap, Roche) and initially validated in vitro using a multidrug-resistant ST175 isolate and representative isolates from major P. aeruginosa clades. In vivo testing included ventilator associated pneumonia by MDR P. aeruginosa in ICU (3-10 sequential samples from 3 patients) and chronic respiratory infection by hypermutable P. aeruginosa in cystic fibrosis (8 sequential samples from a single patient covering a 4-year period). Results from direct sequencing with the enrichment panel were compared with those of whole genome sequencing (WGS) and phenotypic profiling of 10 isolated colonies per sample.
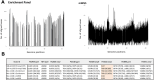
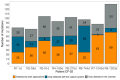
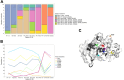
Findings: In vitro assays confirmed the selectivity of the enrichment panel and the correct identification of the vast mutational resistome of ST175, including specific mutations even when introduced in a 1:100 proportion. In vivo performance was at least equivalent to sequencing 10 colonies per sample, including the accurate identification of the sequence types and the basal and acquired mutational resistome. To note, specific resistance mutations, such as those in ampC leading to resistance to novel β-lactams, could be traced even at frequencies of 1%. Moreover, the coselection of mutator populations and antibiotic resistance mutations, predicted in theoretical and in vitro studies, was evidenced in vivo.
Interpretation: This proof-of-concept study demonstrates that resistance genomics of P. aeruginosa can be analysed directly from clinical samples, determining not only a considerable reduction in turnaround time and cost from a diagnostics perspective, but also an unprecedented potency for accurate monitoring of in vivo population dynamics in bacterial infections.
Funding: Instituto de Salud Carlos III, Ministerio de Ciencia e Innovación and Unión Europea-NextGenerationEU.
Keywords: Antimicrobial resistance development; Chronic infections; Cystic fibrosis; Nosocomial infections; Pseudomonas aeruginosa; Resistome.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests AO and RC have participated in educational programs organised by MSD, Pfizer and Shionogi and conducted research studies financed by MSD and Shionogi. JPH has participated in educational programs organised by MSD, Pfizer, Menarini and Angelini and in advisory boards organised by Advanz Pharma, Tillots, and GILEAD.
Figures




References
-
- López-Causapé C., Rojo-Molinero E., Macià M.D., Oliver A. The problems of antibiotic resistance in cystic fibrosis and solutions. Expert Rev Respir Med. 2015;9:73–88. - PubMed
-
- Tacconelli E., Carrara E., Savoldi A., et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18:318–327. - PubMed
-
- Oliver A., Mulet X., López-Causapé C., Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;21-22:41–59. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

