Functionally redundant roles of ID family proteins in spermatogonial stem cells
- PMID: 39332405
- PMCID: PMC11561458
- DOI: 10.1016/j.stemcr.2024.08.011
Functionally redundant roles of ID family proteins in spermatogonial stem cells
Abstract
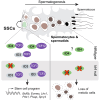
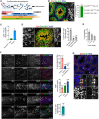
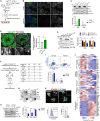
Spermatogonial stem cells (SSCs) are essential for sustained sperm production, but SSC regulatory mechanisms and markers remain poorly defined. Studies have suggested that the Id family transcriptional regulator Id4 is expressed in SSCs and involved in SSC maintenance. Here, we used reporter and knockout models to define the expression and function of Id4 in the adult male germline. Within the spermatogonial pool, Id4 reporter expression and inhibitor of DNA-binding 4 (ID4) protein are found throughout the GFRα1+ fraction, comprising the self-renewing population. However, Id4 deletion is tolerated by adult SSCs while revealing roles in meiotic spermatocytes. Cultures of undifferentiated spermatogonia could be established following Id4 deletion. Importantly, ID4 loss in undifferentiated spermatogonia triggers ID3 upregulation, and both ID proteins associate with transcription factor partner TCF3 in wild-type cells. Combined inhibition of IDs in cultured spermatogonia disrupts the stem cell state and blocks proliferation. Our data therefore demonstrate critical but functionally redundant roles of IDs in SSC function.
Keywords: ID genes; germline; spermatogonial stem cells; transcription factors.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests F.J.R. receives institutional support as a co-investigator and is subcontracted by Peter MacCallum Cancer Centre for an investigator-initiated trial, which receives funding from Sanofi/Regeneron Pharmaceuticals.
Figures

References
-
- Bounpheng M.A., Dimas J.J., Dodds S.G., Christy B.A. Degradation of Id proteins by the ubiquitin-proteasome pathway. FASEB J. 1999;13:2257–2264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

