A living organoid biobank of patients with Crohn's disease reveals molecular subtypes for personalized therapeutics
- PMID: 39332415
- PMCID: PMC11513829
- DOI: 10.1016/j.xcrm.2024.101748
A living organoid biobank of patients with Crohn's disease reveals molecular subtypes for personalized therapeutics
Abstract
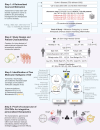
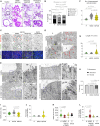
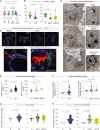
Crohn's disease (CD) is a complex and heterogeneous condition with no perfect preclinical model or cure. To address this, we explore adult stem cell-derived organoids that retain their tissue identity and disease-driving traits. We prospectively create a biobank of CD patient-derived organoid cultures (PDOs) from colonic biopsies of 53 subjects across all clinical subtypes and healthy subjects. Gene expression analyses enabled benchmarking of PDOs as tools for modeling the colonic epithelium in active disease and identified two major molecular subtypes: immune-deficient infectious CD (IDICD) and stress and senescence-induced fibrostenotic CD (S2FCD). Each subtype shows internal consistency in the transcriptome, genome, and phenome. The spectrum of morphometric, phenotypic, and functional changes within the "living biobank" reveals distinct differences between the molecular subtypes. Drug screens reverse subtype-specific phenotypes, suggesting phenotyped-genotyped CD PDOs can bridge basic biology and patient trials by enabling preclinical phase "0" human trials for personalized therapeutics.
Keywords: barrier integrity; host-microbe interaction; inflammatory bowel disease; patient-derived organoids; therapeutics.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests S.D. and P.G. have a patent on the methodology.
Figures



Update of
-
A Living Organoid Biobank of Crohn's Disease Patients Reveals Molecular Subtypes for Personalized Therapeutics.bioRxiv [Preprint]. 2023 Mar 13:2023.03.11.532245. doi: 10.1101/2023.03.11.532245. bioRxiv. 2023. Update in: Cell Rep Med. 2024 Oct 15;5(10):101748. doi: 10.1016/j.xcrm.2024.101748. PMID: 36993763 Free PMC article. Updated. Preprint.
References
-
- Elmentaite R., Ross A.D.B., Roberts K., James K.R., Ortmann D., Gomes T., Nayak K., Tuck L., Pritchard S., Bayraktar O.A., et al. Single-Cell Sequencing of Developing Human Gut Reveals Transcriptional Links to Childhood Crohn's Disease. Dev. Cell. 2020;55:771–783.e5. doi: 10.1016/j.devcel.2020.11.010. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

