Modeling Substitution Rate Evolution across Lineages and Relaxing the Molecular Clock
- PMID: 39332907
- PMCID: PMC11430275
- DOI: 10.1093/gbe/evae199
Modeling Substitution Rate Evolution across Lineages and Relaxing the Molecular Clock
Abstract
Relaxing the molecular clock using models of how substitution rates change across lineages has become essential for addressing evolutionary problems. The diversity of rate evolution models and their implementations are substantial, and studies have demonstrated their impact on divergence time estimates can be as significant as that of calibration information. In this review, we trace the development of rate evolution models from the proposal of the molecular clock concept to the development of sophisticated Bayesian and non-Bayesian methods that handle rate variation in phylogenies. We discuss the various approaches to modeling rate evolution, provide a comprehensive list of available software, and examine the challenges and advancements of the prevalent Bayesian framework, contrasting them to faster non-Bayesian methods. Lastly, we offer insights into potential advancements in the field in the era of big data.
Keywords: model comparison; molecular clock history; molecular dating; rate heterogeneity; rate models; relaxed molecular clock.
© The Author(s) 2024. Published by Oxford University Press on behalf of Society for Molecular Biology and Evolution.
Figures
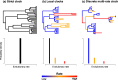
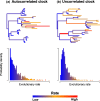
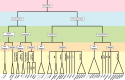
References
-
- Asadollahi M, Boroumand H, Mohammadi S, Mercado-Salas NF, Ahmadzadeh F. Molecular and morphological evidence reveals the presence of the tadpole shrimp Lepidurus cf. couesii (crustacea: Branchiopoda) in Iran. Zool Anz. 2023:306:1–9. 10.1016/j.jcz.2023.06.009. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

