Spatial organization of adenylyl cyclase and its impact on dopamine signaling in neurons
- PMID: 39333071
- PMCID: PMC11436756
- DOI: 10.1038/s41467-024-52575-0
Spatial organization of adenylyl cyclase and its impact on dopamine signaling in neurons
Abstract
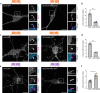
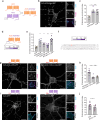
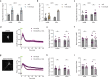
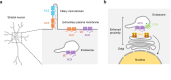
The cAMP cascade is increasingly recognized to transduce physiological effects locally through spatially limited cAMP gradients. However, little is known about how adenylyl cyclase enzymes that initiate cAMP gradients are localized. Here we address this question in physiologically relevant striatal neurons and investigate how AC localization impacts downstream signaling function. We show that the major striatal AC isoforms are differentially sorted between ciliary and extraciliary domains of the plasma membrane, and that one isoform, AC9, is uniquely concentrated in endosomes. We identify key sorting determinants in the N-terminal cytoplasmic domain responsible for isoform-specific localization. We further show that AC9-containing endosomes accumulate activated dopamine receptors and form an elaborately intertwined network with juxtanuclear PKA stores bound to Golgi membranes. Finally, we provide evidence that endosomal localization enables AC9 to selectively elevate PKA activity in the nucleus relative to the cytoplasm. Together, these results reveal a precise spatial landscape of the cAMP cascade in neurons and a key role of AC localization in directing downstream PKA signaling to the nucleus.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Spatial organization of adenylyl cyclase and its impact on dopamine signaling in neurons.bioRxiv [Preprint]. 2023 Dec 6:2023.12.06.570478. doi: 10.1101/2023.12.06.570478. bioRxiv. 2023. Update in: Nat Commun. 2024 Sep 27;15(1):8297. doi: 10.1038/s41467-024-52575-0. PMID: 38106018 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
- ALTF 192-2019/European Molecular Biology Organization (EMBO)
- R35 GM145291/GM/NIGMS NIH HHS/United States
- DA012864/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- DA010711/U.S. Department of Health & Human Services | NIH | National Institute on Drug Abuse (NIDA)
- R29 DA010711/DA/NIDA NIH HHS/United States
- R01 GM060419/GM/NIGMS NIH HHS/United States
- R01 DA012864/DA/NIDA NIH HHS/United States
- R01GM060419/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- GM145291/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
- R37 DA010711/DA/NIDA NIH HHS/United States
- S10 OD017993/OD/NIH HHS/United States
- MH120212/U.S. Department of Health & Human Services | NIH | National Institute of Mental Health (NIMH)
- R01 DA010711/DA/NIDA NIH HHS/United States
- R01 MH120212/MH/NIMH NIH HHS/United States

