USF2 and TFEB compete in regulating lysosomal and autophagy genes
- PMID: 39333072
- PMCID: PMC11436898
- DOI: 10.1038/s41467-024-52600-2
USF2 and TFEB compete in regulating lysosomal and autophagy genes
Abstract
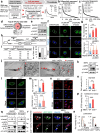
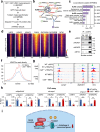
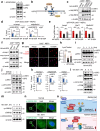
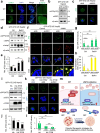
Autophagy, a highly conserved self-digestion process crucial for cellular homeostasis, is triggered by various environmental signals, including nutrient scarcity. The regulation of lysosomal and autophagy-related processes is pivotal to maintaining cellular homeostasis and basal metabolism. The consequences of disrupting or diminishing lysosomal and autophagy systems have been investigated; however, information on the implications of hyperactivating lysosomal and autophagy genes on homeostasis is limited. Here, we present a mechanism of transcriptional repression involving upstream stimulatory factor 2 (USF2), which inhibits lysosomal and autophagy genes under nutrient-rich conditions. We find that USF2, together with HDAC1, binds to the CLEAR motif within lysosomal genes, thereby diminishing histone H3K27 acetylation, restricting chromatin accessibility, and downregulating lysosomal gene expression. Under starvation, USF2 competes with transcription factor EB (TFEB), a master transcriptional activator of lysosomal and autophagy genes, to bind to target gene promoters in a phosphorylation-dependent manner. The GSK3β-mediated phosphorylation of the USF2 S155 site governs USF2 DNA-binding activity, which is involved in lysosomal gene repression. These findings have potential applications in the treatment of protein aggregation-associated diseases, including α1-antitrypsin deficiency. Notably, USF2 repression is a promising therapeutic strategy for lysosomal and autophagy-related diseases.
© 2024. The Author(s).
Conflict of interest statement
A.B. is cofounder of CASMA Therapeutics, Inc., and Advisory board member of NexGeneration Diagnostics and Avilar Therapeutics. The remaining authors declare no competing interests.
Figures



References
-
- Ballabio, A. & Bonifacino, J. S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. cell Biol.21, 101–118 (2020). - PubMed
-
- Parenti, G., Andria, G. & Ballabio, A. Lysosomal storage diseases: from pathophysiology to therapy. Annu. Rev. Med.66, 471–486 (2015). - PubMed
-
- Platt, F. M., d’Azzo, A., Davidson, B. L., Neufeld, E. F. & Tifft, C. J. Lysosomal storage diseases. Nat. Rev. Dis. Prim.4, 27 (2018). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

