The microprotein HDSP promotes gastric cancer progression through activating the MECOM-SPINK1-EGFR signaling axis
- PMID: 39333095
- PMCID: PMC11437185
- DOI: 10.1038/s41467-024-50986-7
The microprotein HDSP promotes gastric cancer progression through activating the MECOM-SPINK1-EGFR signaling axis
Abstract
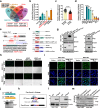
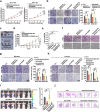
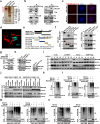
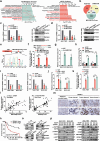
The presence of noncanonical open reading frames within lncRNAs (long non-coding RNAs) suggests their potential for translation, yielding various functional peptides or proteins. However, the existence and specific roles of these products in gastric cancer remain largely unclear. Here we identify the HOXA10-HOXA9-derived small protein (HDSP) in gastric cancer through comprehensive analysis and experimental validation, including mass spectrometry and western blotting. HDSP exhibits high expression and oncogenic roles in gastric cancer. Mechanistically, HDSP blocks TRIM25-mediated ubiquitination and degradation by interacting with MECOM, leading to MECOM accumulation and enhanced SPINK1 transcription-a gene promoting cancer via the EGFR signaling pathway. Furthermore, MECOM fosters HOXA10-HOXA9 transcription, establishing a feedback loop activating SPINK1-EGFR signaling. HDSP knockdown inhibits tumor growth in a PDX (patient-derived xenograft) model, and infusion of an artificially synthesized HDSP peptide as a neoantigen enhances immune cell-mediated anti-tumor efficacy against gastric cancer in vitro and in vivo. These findings propose HDSP as a potential therapeutic target or neoantigen candidate for gastric cancer treatment.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Siegel, R. L., Miller, K. D. & Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin.70, 7–30 (2020). - PubMed
-
- Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin.72, 7–33 (2022). - PubMed
-
- Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin.71, 209–249 (2021). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

