Gut microbiota wellbeing index predicts overall health in a cohort of 1000 infants
- PMID: 39333099
- PMCID: PMC11436675
- DOI: 10.1038/s41467-024-52561-6
Gut microbiota wellbeing index predicts overall health in a cohort of 1000 infants
Abstract
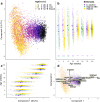
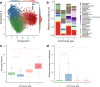
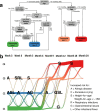
The human gut microbiota is central in regulating all facets of host physiology, and in early life it is thought to influence the host's immune system and metabolism, affecting long-term health. However, longitudinally monitored cohorts with parallel analysis of faecal samples and health data are scarce. In our observational study we describe the gut microbiota development in the first 2 years of life and create a gut microbiota wellbeing index based on the microbiota development and health data in a cohort of nearly 1000 infants using clustering and trajectory modelling. We show that infants' gut microbiota development is highly predictable, following one of five trajectories, dependent on infant exposures, and predictive of later health outcomes. We characterise the natural healthy gut microbiota trajectory and several different dysbiotic trajectories associated with different health outcomes. Bifidobacterium and Bacteroides appear as early keystone organisms, directing microbiota development and consistently predicting positive health outcomes. A microbiota wellbeing index, based on the healthy development trajectory, is predictive of general health over the first 5 years. The results indicate that gut microbiota succession is part of infant physiological development, predictable, and malleable. This information can be utilised to improve the predictions of individual health risks.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

