Histone marks identify novel transcription factors that parse CAR-T subset-of-origin, clinical potential and expansion
- PMID: 39333103
- PMCID: PMC11436946
- DOI: 10.1038/s41467-024-52503-2
Histone marks identify novel transcription factors that parse CAR-T subset-of-origin, clinical potential and expansion
Abstract
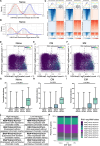
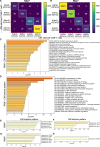
Chimeric antigen receptor-modified T cell (CAR-T) immunotherapy has revolutionised blood cancer treatment. Parsing the genetic underpinnings of T cell quality and CAR-T efficacy is challenging. Transcriptomics inform CAR-T state, but the nature of dynamic transcription during activation hinders identification of transiently or minimally expressed genes, such as transcription factors, and over-emphasises effector and metabolism genes. Here we explore whether analyses of transcriptionally repressive and permissive histone methylation marks describe CAR-T cell functional states and therapeutic potential beyond transcriptomic analyses. Histone mark analyses improve identification of differences between naïve, central memory, and effector memory CD8 + T cell subsets of human origin, and CAR-T derived from these subsets. We find important differences between CAR-T manufactured from central memory cells of healthy donors and of patients. By examining CAR-T products from a clinical trial in lymphoma (NCT01865617), we find a novel association between the activity of the transcription factor KLF7 with in vivo CAR-T accumulation in patients and demonstrate that over-expression of KLF7 increases in vitro CAR-T proliferation and IL-2 production. In conclusion, histone marks provide a rich dataset for identification of functionally relevant genes not apparent by transcriptomics.
© 2024. The Author(s).
Conflict of interest statement
S.F. has filed and received license fees on patents for optimising CAR T cell function, has received research laboratory grants from Bristol Myers Squibb, and has consulted ad hoc for Prescient Therapeutics. E.L.K. receives research support from Juno Therapeutics, a BMS company. A.S.S. is a current employee of Umoja Biopharma. A.V.H. has received research funding from Juno Therapeutics, a Bristol Myers Squibb Company, and Nektar Therapeutics; it has received honoraria from Bristol Myers Squibb. J.G. reports honoraria from EUSA Pharma, JMP, Larvol, and Multerra Bio; serves on scientific advisory boards for Legend Biotech, Janssen, Kite (a Gilead company), and MorphoSys; and receives research funding from Sobi, Juno Therapeutics, Celgene (a BMS company), and Angiocrine Bioscience. S.R.R. is a cofounder and adviser to Lyell Immunopharma; has research funding from and intellectual property licensed to Lyell Immunopharma; was a cofounder of Juno Therapeutics; is an inventor of patents licensed to Juno Therapeutics; and served as an adviser to Juno Therapeutics and Adaptive Biotechnologies. D.G.M. has received research funding from Juno Therapeutics, a Bristol Myers Squibb Company, Celgene, and Kite Pharma, a Gilead company; has served on ad hoc advisory board meetings from Amgen, BMS, Genentech, Gilead, Incyte, Janssen, Legend Biotech, Mustang Bio, MorphoSys, Novartis, Pharmacyclics, and Umoja; has rights to receive royalties from Fred Hutch for patents licensed to Juno Therapeutics; serves on scientific advisory board with stock options and compensations for A2 Biotherapeutics and Navan Technologies. S.H. has filed patent applications on related work. C.J.T. received research funding from Juno Therapeutics/BMS, Nektar Therapeutics, and Nanostring; serves on Scientific Advisory Boards for Caribou Biosciences, T-CURX, Myeloid Therapeutics, ArsenalBio, Cargo Therapeutics, Celgene/BMS Cell Therapy, Differentia Bio, eGlint, and Advesya; is a DSMB member for Kyverna; has served on ad hoc advisory roles/consulting (last 12 months) for Prescient Therapeutics, Century Therapeutics, IGM Biosciences, Abbvie, Boxer Capital, Novartis; holds stock options for Eureka Therapeutics, Caribou Biosciences, Myeloid Therapeutics, ArsenalBio, Cargo Therapeutics, and eGlint; and has the right to receive payment from Fred Hutch Cancer Center as an inventor on patents related to CAR T-cell therapy. The remaining authors declare no competing interest.
Figures






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

