Potent pollen gene regulation by DNA glycosylases in maize
- PMID: 39333110
- PMCID: PMC11436724
- DOI: 10.1038/s41467-024-52620-y
Potent pollen gene regulation by DNA glycosylases in maize
Abstract
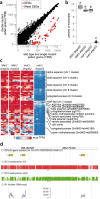
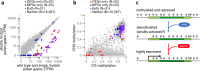
Although DNA methylation primarily represses TEs, it also represses select genes that are methylated in plant body tissues but demethylated by DNA glycosylases (DNGs) in endosperm or pollen. Either one of two DNGs, MATERNAL DEREPRESSION OF R1 (MDR1) or DNG102, is essential for pollen viability in maize. Using single-pollen mRNA sequencing on pollen-segregating mutations in both genes, we identify 58 candidate DNG target genes that account for 11.1% of the wild-type transcriptome but are silent or barely detectable in other tissues. They are unusual in their tendency to lack introns but even more so in their TE-like methylation (teM) in coding DNA. The majority have predicted functions in cell wall modification, and they likely support the rapid tip growth characteristic of pollen tubes. These results suggest a critical role for DNA methylation and demethylation in regulating maize genes with the potential for extremely high expression in pollen but constitutive silencing elsewhere.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Update of
-
Potent pollen gene regulation by DNA glycosylases in maize.bioRxiv [Preprint]. 2024 Jul 19:2024.02.13.580204. doi: 10.1101/2024.02.13.580204. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 27;15(1):8352. doi: 10.1038/s41467-024-52620-y. PMID: 38405940 Free PMC article. Updated. Preprint.
References
-
- Miller, E. C. Development of the pistillate spikelet and fertilization in Zea mays L. J. Agric. Res.18, 12 (1919).
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

