The role of forkhead box M1-methionine adenosyltransferase 2 A/2B axis in liver inflammation and fibrosis
- PMID: 39333125
- PMCID: PMC11436801
- DOI: 10.1038/s41467-024-52527-8
The role of forkhead box M1-methionine adenosyltransferase 2 A/2B axis in liver inflammation and fibrosis
Abstract
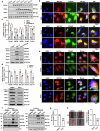
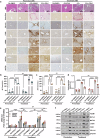
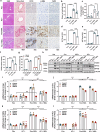
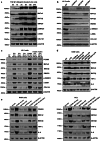
Methionine adenosyltransferase 2 A (MAT2A) and MAT2B are essential for hepatic stellate cells (HSCs) activation. Forkhead box M1 (FOXM1) transgenic mice develop liver inflammation and fibrosis. Here we examine if they crosstalk in male mice. We found FOXM1/MAT2A/2B are upregulated after bile duct ligation (BDL) and carbon tetrachloride (CCl4) treatment in hepatocytes, HSCs and Kupffer cells (KCs). FDI-6, a FOXM1 inhibitor, attenuates the development and reverses the progression of CCl4-induced fibrosis while lowering the expression of FOXM1/MAT2A/2B, which exert reciprocal positive regulation on each other transcriptionally. Knocking down any of them lowers HSCs and KCs activation. Deletion of FOXM1 in hepatocytes, HSCs, and KCs protects from BDL-mediated inflammation and fibrosis comparably. Interestingly, HSCs from Foxm1Hep-/-, hepatocytes from Foxm1HSC-/-, and HSCs and hepatocytes from Foxm1KC-/- have lower FOXM1/MAT2A/2B after BDL. This may be partly due to transfer of extracellular vesicles between different cell types. Altogether, FOXM1/MAT2A/MAT2B axis drives liver inflammation and fibrosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Friedman, S. L. Liver fibrosis – from bench to bedside. J. Hepatol.38, S38–S53 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

