A basally active cGAS-STING pathway limits SARS-CoV-2 replication in a subset of ACE2 positive airway cell models
- PMID: 39333139
- PMCID: PMC11437049
- DOI: 10.1038/s41467-024-52803-7
A basally active cGAS-STING pathway limits SARS-CoV-2 replication in a subset of ACE2 positive airway cell models
Abstract
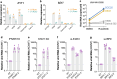
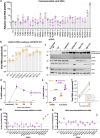
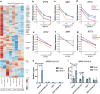
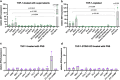
Host factors that define the cellular tropism of SARS-CoV-2 beyond the cognate ACE2 receptor are poorly defined. Here we report that SARS-CoV-2 replication is restricted at a post-entry step in a number of ACE2-positive airway-derived cell lines due to tonic activation of the cGAS-STING pathway mediated by mitochondrial DNA leakage and naturally occurring cGAS and STING variants. Genetic and pharmacological inhibition of the cGAS-STING and type I/III IFN pathways as well as ACE2 overexpression overcome these blocks. SARS-CoV-2 replication in STING knockout cell lines and primary airway cultures induces ISG expression but only in uninfected bystander cells, demonstrating efficient antagonism of the type I/III IFN-pathway in productively infected cells. Pharmacological inhibition of STING in primary airway cells enhances SARS-CoV-2 replication and reduces virus-induced innate immune activation. Together, our study highlights that tonic activation of the cGAS-STING and IFN pathways can impact SARS-CoV-2 cellular tropism in a manner dependent on ACE2 expression levels.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Update of
-
A basally active cGAS-STING pathway limits SARS-CoV-2 replication in a subset of ACE2 positive airway cell models.bioRxiv [Preprint]. 2024 Sep 6:2024.01.07.574522. doi: 10.1101/2024.01.07.574522. bioRxiv. 2024. Update in: Nat Commun. 2024 Sep 27;15(1):8394. doi: 10.1038/s41467-024-52803-7. PMID: 38260460 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- T32CA009547-34/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- T32 DK007130/DK/NIDDK NIH HHS/United States
- R56 AI167285/AI/NIAID NIH HHS/United States
- AI167695/Division of Intramural Research, National Institute of Allergy and Infectious Diseases (Division of Intramural Research of the NIAID)
- F31 AI167695/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

