SURF2 is a MDM2 antagonist in triggering the nucleolar stress response
- PMID: 39333141
- PMCID: PMC11436901
- DOI: 10.1038/s41467-024-52659-x
SURF2 is a MDM2 antagonist in triggering the nucleolar stress response
Abstract
Cancer cells rely on high ribosome production to sustain their proliferation rate. Many chemotherapies impede ribosome production which is perceived by cells as "nucleolar stress" (NS), triggering p53-dependent and independent pathways leading to cell cycle arrest and/or apoptosis. The 5S ribonucleoprotein (RNP) particle, a sub-ribosomal particle, is instrumental to NS response. Upon ribosome assembly defects, the 5S RNP accumulates as free form. This free form is able to sequester and inhibit MDM2, thus promoting p53 stabilization. To investigate how cancer cells can resist to NS, here we purify free 5S RNP and uncover an interaction partner, SURF2. Functional characterization of SURF2 shows that its depletion increases cellular sensitivity to NS, while its overexpression promotes their resistance to it. Consistently, SURF2 is overexpressed in many cancers and its expression level is an independent marker of prognosis for adrenocortical cancer. Our data demonstrate that SURF2 buffers free 5S RNP particles, and can modulate their activity, paving the way for the research of new molecules that can finely tune the response to nucleolar stress in the framework of cancer therapies.
© 2024. The Author(s).
Conflict of interest statement
S.T., S.L., V.M., C.P.C., and P.E.G. have filed a patent application on the targeting of SURF2 in cancer, therapies and ribosomopathies treatments. The remaining authors declare no competing interests.
Figures
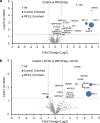


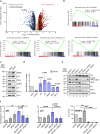
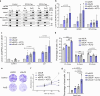
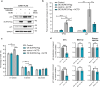
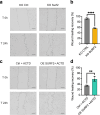

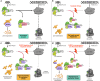
References
-
- Derenzini, M. et al. Nucleolar size indicates the rapidity of cell proliferation in cancer tissues. J. Pathol.191, 181–186 (2000). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

