Vitessce: integrative visualization of multimodal and spatially resolved single-cell data
- PMID: 39333268
- PMCID: PMC11725496
- DOI: 10.1038/s41592-024-02436-x
Vitessce: integrative visualization of multimodal and spatially resolved single-cell data
Abstract
Multiomics technologies with single-cell and spatial resolution make it possible to measure thousands of features across millions of cells. However, visual analysis of high-dimensional transcriptomic, proteomic, genome-mapped and imaging data types simultaneously remains a challenge. Here we describe Vitessce, an interactive web-based visualization framework for exploration of multimodal and spatially resolved single-cell data. We demonstrate integrative visualization of millions of data points, including cell-type annotations, gene expression quantities, spatially resolved transcripts and cell segmentations, across multiple coordinated views. The open-source software is available at http://vitessce.io .
© 2024. The Author(s).
Conflict of interest statement
Competing interests: N.G. is a co-founder and equity owner of Datavisyn. P.V.K. serves on the Scientific Advisory Board to Celsius Therapeutics, Inc. and Biomage, Inc. The other authors declare no competing interests.
Figures



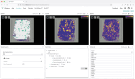
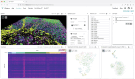

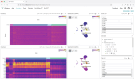
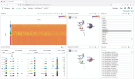
References
-
- Samatova, N. F., Breimyer, P., Hendrix, W., Schmidt, M. C. & Rhyne, T. An outlook into ultra-scale visualization of large-scale biological data. In 2008 Workshop on Ultrascale Visualization 29–39 (IEEE, 2008).
-
- Garrison, L. A., Kolesar, I., Viola, I., Hauser, H. & Bruckner, S. Trends and opportunities in visualization for physiology: a multiscale overview. Comput. Graph. Forum41, 609–643 (2022).
-
- Megill, C., et al. Cellxgene: a performant, scalable exploration platform for high dimensional sparse matrices. Preprint at bioRxiv10.1101/2021.04.05.438318 (2021).
MeSH terms
Grants and funding
- DGE1745303/National Science Foundation (NSF)
- U54 DK120058/DK/NIDDK NIH HHS/United States
- T15LM007092/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2OD026671/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2 OD026671/OD/NIH HHS/United States
- T32 HG002295/HG/NHGRI NIH HHS/United States
- OT2OD033758/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- 2P41GM103391/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54DK120058/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- T32HG002295/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- T15 LM007092/LM/NLM NIH HHS/United States
- R33CA263666/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P41 GM103391/GM/NIGMS NIH HHS/United States
- R33 CA263666/CA/NCI NIH HHS/United States
- OT2 OD026677/OD/NIH HHS/United States
- OT2 OD033758/OD/NIH HHS/United States
- CBET1828299/National Science Foundation (NSF)
LinkOut - more resources
Full Text Sources

