An in silico design method of a peptide bioreceptor for cortisol using molecular modelling techniques
- PMID: 39333310
- PMCID: PMC11436820
- DOI: 10.1038/s41598-024-73044-0
An in silico design method of a peptide bioreceptor for cortisol using molecular modelling techniques
Abstract
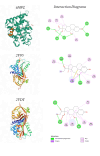
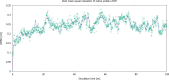
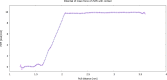
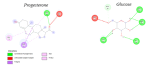
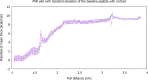
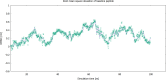
Cortisol is established as a reliable biomarker for stress prompting intensified research in developing wearable sensors to detect it via eccrine sweat. Since cortisol is present in sweat in trace quantities, typically 8-140 ng/mL, developing such biosensors necessitates the design of bioreceptors with appropriate sensitivity and selectivity. In this work, we present a systematic biomimetic methodology and a semi-automated high-throughput screening tool which enables rapid selection of bioreceptors as compared to ab initio design of peptides via computational peptidology. Candidate proteins from databases are selected via molecular docking and ranked according to their binding affinities by conducting automated AutoDock Vina scoring simulations. These candidate proteins are then validated via full atomistic steered molecular dynamics computations including umbrella sampling to estimate the potential of mean force using GROMACS version 2022.6. These explicit molecular dynamic calculations are carried out in an eccrine sweat environment taking into consideration the protein dynamics and solvent effects. Subsequently, we present a candidate baseline peptide bioreceptor selected as a contiguous sequence of amino acids from the selected protein binding pocket favourably interacting with the target ligand (i.e., cortisol) from the active binding site of the proteins and maintaining its tertiary structure. A unique cysteine residue introduced at the N-terminus allows orientation-specific surface immobilization of the peptide onto the gold electrodes and to ensure exposure of the binding site. Comparative binding affinity simulations of this peptide with the target ligand along with commonly interfering species e.g., progesterone, testosterone and glucose are also presented to demonstrate the validity of this proposed peptide as a candidate baseline bioreceptor for future cortisol biosensor development.
Keywords: Computational peptidology; Cortisol bioreceptor; De novo peptide design; High-throughput screening; Molecular docking; Molecular dynamics; Umbrella sampling.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Bandodkar, A. J., Ghaffari, R. & Rogers, J. A. Don’t sweat it: The Quest for wearable stress sensors. Matter. 2, 795–797 (2020). - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

