Telomere-to-telomere assemblies of cattle and sheep Y-chromosomes uncover divergent structure and gene content
- PMID: 39333471
- PMCID: PMC11436988
- DOI: 10.1038/s41467-024-52384-5
Telomere-to-telomere assemblies of cattle and sheep Y-chromosomes uncover divergent structure and gene content
Abstract
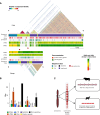
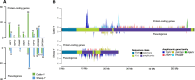
Reference genomes of cattle and sheep have lacked contiguous assemblies of the sex-determining Y chromosome. Here, we assemble complete and gapless telomere to telomere (T2T) Y chromosomes for these species. We find that the pseudo-autosomal regions are similar in length, but the total chromosome size is substantially different, with the cattle Y more than twice the length of the sheep Y. The length disparity is accounted for by expanded ampliconic region in cattle. The genic amplification in cattle contrasts with pseudogenization in sheep suggesting opposite evolutionary mechanisms since their divergence 19MYA. The centromeres also differ dramatically despite the close relationship between these species at the overall genome sequence level. These Y chromosomes have been added to the current reference assemblies in GenBank opening new opportunities for the study of evolution and variation while supporting efforts to improve sustainability in these important livestock species that generally use sire-driven genetic improvement strategies.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
T.A.O. and S.K. have received travel funds to speak at events hosted by Oxford Nanopore Technologies. The remaining authors declare no competing interests.
Figures

Update of
-
The first complete T2T Assemblies of Cattle and Sheep Y-Chromosomes uncover remarkable divergence in structure and gene content.Res Sq [Preprint]. 2024 Apr 3:rs.3.rs-4033388. doi: 10.21203/rs.3.rs-4033388/v1. Res Sq. 2024. Update in: Nat Commun. 2024 Sep 27;15(1):8277. doi: 10.1038/s41467-024-52384-5. PMID: 38712074 Free PMC article. Updated. Preprint.
References
-
- Luo, Z.-X., Yuan, C.-X., Meng, Q.-J. & Ji, Q. A Jurassic eutherian mammal and divergence of marsupials and placentals. Nature10.1038/nature10291 (2011). - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
- USDA-NIFA-2021-67016-33416/United States Department of Agriculture | National Institute of Food and Agriculture (NIFA)
- IDA01566/United States Department of Agriculture | National Institute of Food and Agriculture (NIFA)
- 3040-31000-104-000-D/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
- 8042-31000-112-000-D/United States Department of Agriculture | Agricultural Research Service (USDA Agricultural Research Service)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

