Scar matrix drives Piezo1 mediated stromal inflammation leading to placenta accreta spectrum
- PMID: 39333481
- PMCID: PMC11436960
- DOI: 10.1038/s41467-024-52351-0
Scar matrix drives Piezo1 mediated stromal inflammation leading to placenta accreta spectrum
Abstract
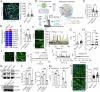
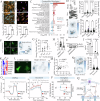
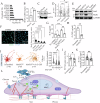
Scar tissue formation is a hallmark of wound repair in adults and can chronically affect tissue architecture and function. To understand the general phenomena, we sought to explore scar-driven imbalance in tissue homeostasis caused by a common, and standardized surgical procedure, the uterine scar due to cesarean surgery. Deep uterine scar is associated with a rapidly increasing condition in pregnant women, placenta accreta spectrum (PAS), characterized by aggressive trophoblast invasion into the uterus, frequently necessitating hysterectomy at parturition. We created a model of uterine scar, recapitulating PAS-like invasive phenotype, showing that scar matrix activates mechanosensitive ion channel, Piezo1, through glycolysis-fueled cellular contraction. Piezo1 activation increases intracellular calcium activity and Protein kinase C activation, leading to NF-κB nuclear translocation, and MafG stabilization. This inflammatory transformation of decidua leads to production of IL-8 and G-CSF, chemotactically recruiting invading trophoblasts towards scar, initiating PAS. Our study demonstrates aberrant mechanics of scar disturbs stroma-epithelia homeostasis in placentation, with implications in cancer dissemination.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
- 5R37CA248161/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- K99 HD105973/HD/NICHD NIH HHS/United States
- R01 HD112424/HD/NICHD NIH HHS/United States
- 1R01HD112424/U.S. Department of Health & Human Services | NIH | Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)
- R37 CA248161/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

