Impact of pneumococcal conjugate vaccines on invasive pneumococcal disease-causing lineages among South African children
- PMID: 39333488
- PMCID: PMC11436952
- DOI: 10.1038/s41467-024-52459-3
Impact of pneumococcal conjugate vaccines on invasive pneumococcal disease-causing lineages among South African children
Abstract
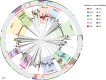
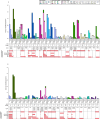
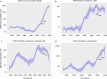
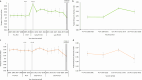
Invasive pneumococcal disease (IPD) due to non-vaccine serotypes after the introduction of pneumococcal conjugate vaccines (PCV) remains a global concern. This study used pathogen genomics to evaluate changes in invasive pneumococcal lineages before, during and after vaccine introduction in South Africa. We included genomes (N = 3104) of IPD isolates from individuals aged <18 years (2005-20), spanning four periods: pre-PCV, PCV7, early-PCV13, and late-PCV13. Significant incidence reductions occurred among vaccine-type lineages in the late-PCV13 period compared to the pre-PCV period. However, some vaccine-type lineages continued to cause invasive disease and showed increasing effective population size trends in the post-PCV era. A significant increase in lineage diversity was observed from the PCV7 period to the early-PCV13 period (Simpson's diversity index: 0.954, 95% confidence interval 0.948-0.961 vs 0.965, 0.962-0.969) supporting intervention-driven population structure perturbation. Increases in the prevalence of penicillin, erythromycin, and multidrug resistance were observed among non-vaccine serotypes in the late-PCV13 period compared to the pre-PCV period. In this work we highlight the importance of continued genomic surveillance to monitor disease-causing lineages post vaccination to support policy-making and future vaccine designs and considerations.
© 2024. The Author(s).
Conflict of interest statement
A.v.G. reports grants from NIH (training grant), US CDC, SEQAFRICA/Fleming Fund, and chairperson of the NITAG for SA (NAGI: National Advisory Group for Immunisation). C.C. reports grants from the US CDC, BMGF, Wellcome, SA MRC, Sanofi Pasteur, and PATH outside the submitted work. J.K. reports support for meetings and travel from NIH, outside the submitted work. R.A.G. reports a grant from Pfizer outside the submitted work. S.WL reports the Robert Austrian Research Award, sponsored by Pfizer, outside the submitted work. S.M. reports EMGuidance Technologies educational event/presentation, non-paid consultant for MSD, and non-paid educational lectures for Sanofi, outside the submitted work. V.Q. reports MSD support for meetings and travel, outside the submitted work. S.W. reports grants from US CDC and BMGF, outside the submitted work. The remaining authors declare no competing interest.
Figures

References
-
- Savulescu, C. et al. Effect of high-valency pneumococcal conjugate vaccines on invasive pneumococcal disease in children in SpIDnet countries: an observational multicentre study. Lancet Respir. Med.5, 648–656 (2017). - PubMed
-
- Madhi, S. A., Cohen, C. & von Gottberg, A. Introduction of pneumococcal conjugate vaccine into the public immunization program in South Africa: translating research into policy. Vaccine30, C21–C27 (2012). - PubMed
-
- von Gottberg, A. et al. Effects of vaccination on invasive pneumococcal disease in South Africa. Vaccine371, 4200–4208 (2014). - PubMed
-
- von Gottberg, A. et al. Long-Term Impact of Pneumococcal Conjugate Vaccines from National, Active, Laboratory-Based Surveillance of Invasive Pneumococcal Disease, South Africa. 10.2139/SSRN.4519527 (2005–2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

