Altered molecular and cellular mechanisms in KIF5A-associated neurodegenerative or neurodevelopmental disorders
- PMID: 39333504
- PMCID: PMC11437142
- DOI: 10.1038/s41419-024-07096-5
Altered molecular and cellular mechanisms in KIF5A-associated neurodegenerative or neurodevelopmental disorders
Abstract
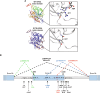
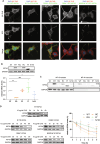
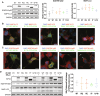
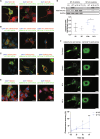
Mutations targeting distinct domains of the neuron-specific kinesin KIF5A associate with different neurodegenerative/neurodevelopmental disorders, but the molecular bases of this clinical heterogeneity are unknown. We characterised five key mutants covering the whole spectrum of KIF5A-related phenotypes: spastic paraplegia (SPG, R17Q and R280C), Charcot-Marie-Tooth disease (CMT, R864*), amyotrophic lateral sclerosis (ALS, N999Vfs*40), and neonatal intractable myoclonus (NEIMY, C975Vfs*73) KIF5A mutants. CMT-R864*-KIF5A and ALS-N999Vfs*40-KIF5A showed impaired autoinhibition and peripheral localisation accompanied by altered mitochondrial distribution, suggesting transport competence disruption. ALS-N999Vfs*40-KIF5A formed SQSTM1/p62-positive inclusions sequestering WT-KIF5A, indicating a gain of toxic function. SPG-R17Q-KIF5A and ALS-N999Vfs*40-KIF5A evidenced a shorter half-life compared to WT-KIF5A, and proteasomal blockage determined their accumulation into detergent-insoluble inclusions. Interestingly, SPG-R280C-KIF5A and ALS-N999Vfs*40-KIF5A both competed for degradation with proteasomal substrates. Finally, NEIMY-C975Vfs*73-KIF5A displayed a similar, but more severe aberrant behaviour compared to ALS-N999Vfs*40-KIF5A; these two mutants share an abnormal tail but cause disorders on the opposite end of KIF5A-linked phenotypic spectrum. Thus, our observations support the pathogenicity of novel KIF5A mutants, highlight abnormalities of recurrent variants, and demonstrate that both unique and shared mechanisms underpin KIF5A-related diseases.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
- GGP19128/Fondazione Telethon (Telethon Foundation)
- 23236/AFM-Téléthon (French Muscular Dystrophy Association)
- PRIN n. 2022EFLFL8/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- PRIN n. P2022B5J32/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- Scientific Exchange Grant n. 9643/European Molecular Biology Organization (EMBO)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

