Split crRNA with CRISPR-Cas12a enabling highly sensitive and multiplexed detection of RNA and DNA
- PMID: 39333528
- PMCID: PMC11436650
- DOI: 10.1038/s41467-024-52691-x
Split crRNA with CRISPR-Cas12a enabling highly sensitive and multiplexed detection of RNA and DNA
Abstract
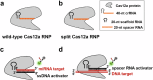
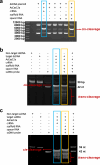
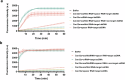
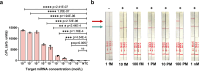
The CRISPR-Cas12a system has revolutionized nucleic acid testing (NAT) with its rapid and precise capabilities, yet it traditionally required RNA pre-amplification. Here we develop rapid fluorescence and lateral flow NAT assays utilizing a split Cas12a system (SCas12a), consisting of a Cas12a enzyme and a split crRNA. The SCas12a assay enables highly sensitive, amplification-free, and multiplexed detection of miRNAs and long RNAs without complex secondary structures. It can differentiate between mature miRNA and its precursor (pre-miRNA), a critical distinction for precise biomarker identification and cancer progression monitoring. The system's specificity is further highlighted by its ability to detect DNA and miRNA point mutations. Notably, the SCas12a system can quantify the miR-21 biomarker in plasma from cervical cancer patients and, when combined with RPA, detect HPV at attomole levels in clinical samples. Together, our work presents a simple and cost-effective SCas12a-based NAT platform for various diagnostic settings.
© 2024. The Author(s).
Conflict of interest statement
Y.L. is a professor of bioscience at Hubei university, and a scientific advisor to BravoVax. The regents of Wuhan Polytechnic University and BravoVax have two patents (application number 2023106008815.7 and 202310602086.4) pending for CRISPR-Cas12a detection technologies on which professor Y.L. and J.Q. are inventors. The remaining authors declare no competing interests.
Figures

References
-
- Kubista, M. et al. The real-time polymerase chain reaction. Mol. Aspects. Med.27, 95–125 (2006). - PubMed
-
- Miller, J. R. & Andre, R. Quantitative polymerase chain reaction. Br. J. Hosp. Med.75, C188–C192 (2014). - PubMed
-
- Li, L. et al. HOLMESv2: A CRISPR-Cas12b-Assisted Platform for Nucleic Acid Detection and DNA Methylation Quantitation. ACS. Synth. Biol.8, 2228–2237 (2019). - PubMed

