piRNAs are regulators of metabolic reprogramming in stem cells
- PMID: 39333531
- PMCID: PMC11437085
- DOI: 10.1038/s41467-024-52709-4
piRNAs are regulators of metabolic reprogramming in stem cells
Abstract
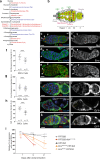
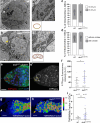
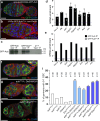
Stem cells preferentially use glycolysis instead of oxidative phosphorylation and this metabolic rewiring plays an instructive role in their fate; however, the underlying molecular mechanisms remain largely unexplored. PIWI-interacting RNAs (piRNAs) and PIWI proteins have essential functions in a range of adult stem cells across species. Here, we show that piRNAs and the PIWI protein Aubergine (Aub) are instrumental in activating glycolysis in Drosophila female germline stem cells (GSCs). Higher glycolysis is required for GSC self-renewal and aub loss-of-function induces a metabolic switch in GSCs leading to their differentiation. Aub directly binds glycolytic mRNAs and Enolase mRNA regulation by Aub depends on its 5'UTR. Furthermore, mutations of a piRNA target site in Enolase 5'UTR lead to GSC loss. These data reveal an Aub/piRNA function in translational activation of glycolytic mRNAs in GSCs, and pinpoint a mechanism of regulation of metabolic reprogramming in stem cells based on small RNAs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- ANR-15-CE12-0019-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-19-CE12-0031/Agence Nationale de la Recherche (French National Research Agency)
- ANR-21-CE12-0035-01/Agence Nationale de la Recherche (French National Research Agency)
- France 2030/Agence Nationale de la Recherche (French National Research Agency)
- Equipe FRM EQU202303016322/Fondation pour la Recherche Médicale (Foundation for Medical Research in France)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

