Identification of potent and orally efficacious phosphodiesterase inhibitors in Cryptosporidium parvum-infected immunocompromised male mice
- PMID: 39333545
- PMCID: PMC11436873
- DOI: 10.1038/s41467-024-52658-y
Identification of potent and orally efficacious phosphodiesterase inhibitors in Cryptosporidium parvum-infected immunocompromised male mice
Abstract
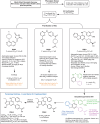
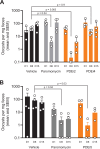
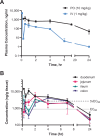
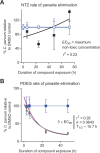
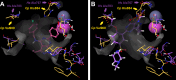
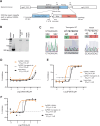
Cryptosporidium parvum and C. hominis are parasites that cause life-threatening diarrhea in children and immunocompromised people. There is only one approved treatment that is modestly effective for children and ineffective for AIDS patients. Here, screening 278 compounds from the Merck KGaA, Darmstadt, Germany collection and accelerated follow-up enabled by prior investigation of the compounds identifies a series of pyrazolopyrimidine human phosphodiesterase (PDE)-V (hPDE-V) inhibitors with potent anticryptosporidial activity and efficacy following oral administration in C. parvum-infected male mice. The lead compounds affect parasite host cell egress, inhibit both C. parvum and C. hominis, work rapidly, and have minimal off-target effects in a safety screening panel. Interestingly, the hPDE-V inhibitors sildenafil and the 4-aminoquinoline compound 7a do not affect Cryptosporidium. C. parvum expresses one PDE (CpPDE1) continuously during asexual growth, the inhibited life stage. According to homology modeling and docking, the lead compounds interact with CpPDE1. Bulkier amino acids (Val900 and His884) in the CpPDE1 active site replace alanines in hPDE-V and block sildenafil binding. Supporting this, sildenafil kills a CRISPR-engineered Cryptosporidium CpPDE1 V900A mutant. The CpPDE1 mutation also alters parasite susceptibility to pyrazolopyrimidines. CpPDE1 is therefore a validated pyrazolopyrimidine molecular target to exploit for target-based optimization for improved anticryptosporidial development.
© 2024. The Author(s).
Conflict of interest statement
C.D.H., M.J.M., and M.G. are inventors on a pending patent application jointly filed by the University of Vermont and St. Louis University that covers use of pyrazolopyrimidines as anti-infective agents (PCT Application No. 63/504,716 filed May 26, 2023). All other authors declare no competing interests.
Figures
References
-
- Hlavsa, M. C. et al. Surveillance for waterborne disease outbreaks and other health events associated with recreational water–United States, 2007–2008. MMWR Surveill. Summ.60, 1–32 (2011). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

