Length-scale study in deep learning prediction for non-small cell lung cancer brain metastasis
- PMID: 39333630
- PMCID: PMC11436900
- DOI: 10.1038/s41598-024-73428-2
Length-scale study in deep learning prediction for non-small cell lung cancer brain metastasis
Abstract
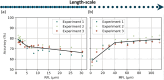
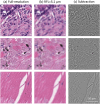
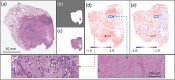
Deep learning-assisted digital pathology has demonstrated the potential to profoundly impact clinical practice, even surpassing human pathologists in performance. However, as deep neural network (DNN) architectures grow in size and complexity, their explainability decreases, posing challenges in interpreting pathology features for broader clinical insights into physiological diseases. To better assess the interpretability of digital microscopic images and guide future microscopic system design, we developed a novel method to study the predictive feature length-scale that underpins a DNN's predictive power. We applied this method to analyze a DNN's capability in predicting brain metastasis from early-stage non-small-cell lung cancer biopsy slides. This study quantifies DNN's attention for brain metastasis prediction, targeting features at both the cellular scale and tissue scale in H&E-stained histological whole slide images. At the cellular scale, the predictive power of DNNs progressively increases with higher resolution and significantly decreases when the resolvable feature length exceeds 5 microns. Additionally, DNN uses more macro-scale features associated with tissue architecture and is optimized when assessing visual fields greater than 41 microns. Our study computes the length-scale requirements for optimal DNN learning on digital whole-slide microscopic images, holding the promise to guide future optical microscope designs in pathology applications and facilitating downstream deep learning analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
MeSH terms
Grants and funding
- 13520296/Sensing to Intelligence (S2I)
- 13520296/Sensing to Intelligence (S2I)
- 13520296/Sensing to Intelligence (S2I)
- 13520296/Sensing to Intelligence (S2I)
- HMRI-15-09-01/Heritage Research Institute for the Advancement of Medicine and Science at Caltech
- HMRI-15-09-01/Heritage Research Institute for the Advancement of Medicine and Science at Caltech
- HMRI-15-09-01/Heritage Research Institute for the Advancement of Medicine and Science at Caltech
- HMRI-15-09-01/Heritage Research Institute for the Advancement of Medicine and Science at Caltech
- U01CA233363/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

