An Enhanced Retroviral Vector for Efficient Genetic Manipulation and Selection in Mammalian Cells
- PMID: 39334897
- PMCID: PMC11430422
- DOI: 10.3390/biom14091131
An Enhanced Retroviral Vector for Efficient Genetic Manipulation and Selection in Mammalian Cells
Abstract
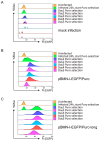
Introducing genetic material into hard-to-transfect mammalian cell lines and primary cells is often best achieved through retroviral infection. An ideal retroviral vector should offer a compact, selectable, and screenable marker while maximizing transgene delivery capacity. However, a previously published retroviral vector featuring an EGFP/Puromycin fusion protein failed to meet these criteria in our experiments. We encountered issues such as low infection efficiency, weak EGFP fluorescence, and selection against infected cells. To address these shortcomings, we developed a novel retroviral vector based on the Moloney murine leukemia virus. This vector includes a compact bifunctional EGFP and Puromycin resistance cassette connected by a 2A peptide. Our extensively tested vector demonstrated superior EGFP expression, efficient Puromycin selection, and no growth penalty in infected cells compared with the earlier design. These benefits were consistent across multiple mammalian cell types, underscoring the versatility of our vector. In summary, our enhanced retroviral vector offers a robust solution for efficient infection, reliable detection, and effective selection in mammalian cells. Its improved performance and compact design make it an ideal choice for a wide range of applications involving precise genetic manipulation and characterization in cell-based studies.
Keywords: B cells; EGFP; FACS; Puromycin; T2A; fusion protein; infection; linker; retrovirus.
Conflict of interest statement
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Figures



References
-
- Miller A.D. Development and applications of retroviral vectors. In: Coffin J.M., Hughes S.H., Varmus H.E., editors. Retroviruses. Cold Spring Harbor; New York, NY, USA: 1997. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

