Lactate Suppresses Growth of Esophageal Adenocarcinoma Patient-Derived Organoids through Alterations in Tumor NADH/NAD+ Redox State
- PMID: 39334961
- PMCID: PMC11430592
- DOI: 10.3390/biom14091195
Lactate Suppresses Growth of Esophageal Adenocarcinoma Patient-Derived Organoids through Alterations in Tumor NADH/NAD+ Redox State
Abstract
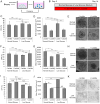
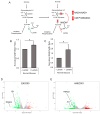
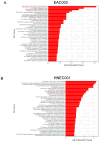
Barrett's esophagus (BE) is a common precancerous lesion that can progress to esophageal adenocarcinoma (EAC). There are significant alterations in the esophageal microbiome in the progression from healthy esophagus to BE to EAC, including an increased abundance of a variety of lactate-producing bacteria and an increase of lactate in the tumor microenvironment, as predicted by metabolic modeling. The role of bacterial lactate in EAC is unknown. Here, we utilize patient-derived organoid (PDO) models of EAC and demonstrate that lactate inhibits the growth and proliferation of EAC PDOs through alterations in the tumor NADH/NAD+ redox state. Further RNA sequencing of EAC PDOs identifies ID1 and RSAD2 as potential regulatory molecules crucial in mediating lactate's ability to suppress glycolysis and proliferation. Gene ontology analysis also identifies the activation of inflammatory and immunological pathways in addition to alterations in the metabolic pathways in EAC PDOs exposed to lactate, suggesting a multi-faceted role for lactate in the pathogenesis of EAC.
Keywords: esophageal adenocarcinoma; lactate; tumor microenvironment.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

