Analysis of Antibiotic Resistance Genes (ARGs) across Diverse Bacterial Species in Shrimp Aquaculture
- PMID: 39334999
- PMCID: PMC11429446
- DOI: 10.3390/antibiotics13090825
Analysis of Antibiotic Resistance Genes (ARGs) across Diverse Bacterial Species in Shrimp Aquaculture
Abstract
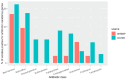
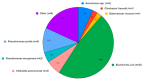
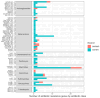
There is little information available on antibiotic resistance (ABR) within shrimp aquaculture environments. The aim of this study was to investigate the presence of antibiotic resistance genes (ARGs) in shrimp farming operations in Atacames, Ecuador. Water samples (n = 162) and shrimp samples (n = 54) were collected from three shrimp farming operations. Samples were cultured and a subset of isolates that grew in the presence of ceftriaxone, a third-generation cephalosporin, were analyzed using whole-genome sequencing (WGS). Among the sequenced isolates (n = 44), 73% of the isolates contained at least one ARG and the average number of ARGs per isolate was two, with a median of 3.5 ARGs. Antibiotic resistance genes that confer resistance to the β-lactam class of antibiotics were observed in 65% of the sequenced isolates from water (20/31) and 54% of the isolates from shrimp (7/13). We identified 61 different ARGs across the 44 sequenced isolates, which conferred resistance to nine antibiotic classes. Over half of all sequenced isolates (59%, n = 26) carried ARGs that confer resistance to more than one class of antibiotics. ARGs for certain antibiotic classes were more common, including beta-lactams (26 ARGs); aminoglycosides (11 ARGs); chloramphenicol (three ARGs); and trimethoprim (four ARGs). Sequenced isolates consisted of a diverse array of bacterial orders and species, including Escherichia coli (48%), Klebsiella pneumoniae (7%), Aeromonadales (7%), Pseudomonadales (16%), Enterobacter cloacae (2%), and Citrobacter freundii (2%). Many ARGs were shared across diverse species, underscoring the risk of horizontal gene transfer in these environments. This study indicated the widespread presence of extended-spectrum β-lactamase (ESBL) genes in shrimp aquaculture, including blaCTX-M, blaSHV, and blaTEM genes. Increased antibiotic resistance surveillance of shrimp farms and identification of aquaculture operation-level risk factors, such as antibiotic use, will likely be important for mitigating the spread of ARGs of clinical significance.
Keywords: 3GCR; AMR; E. coli; aquaculture; ceftriaxone antibiotic resistance; genomic analysis; multi-drug resistance; public health; shrimp.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- World Health Organization . Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report 2022. World Health Organization; Geneva, Switzerland: 2022.
-
- Hossain A., Habibullah-Al-Mamun M., Nagano I., Masunaga S., Kitazawa D., Matsuda H. Antibiotics, Antibiotic-Resistant Bacteria, and Resistance Genes in Aquaculture: Risks, Current Concern, and Future Thinking. Environ. Sci. Pollut. Res. Int. 2022;29:11054–11075. doi: 10.1007/s11356-021-17825-4. - DOI - PubMed
-
- Schar D., Sommanustweechai A., Laxminarayan R., Tangcharoensathien V. Surveillance of Antimicrobial Consumption in Animal Production Sectors of Low- and Middle-Income Countries: Optimizing Use and Addressing Antimicrobial Resistance. PLoS Med. 2018;15:e1002521. doi: 10.1371/journal.pmed.1002521. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

