Protein Structure Inspired Discovery of a Novel Inducer of Anoikis in Human Melanoma
- PMID: 39335149
- PMCID: PMC11429909
- DOI: 10.3390/cancers16183177
Protein Structure Inspired Discovery of a Novel Inducer of Anoikis in Human Melanoma
Abstract
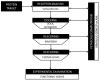
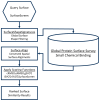
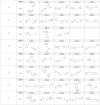
Drug discovery historically starts with an established function, either that of compounds or proteins. This can hamper discovery of novel therapeutics. As structure determines function, we hypothesized that unique 3D protein structures constitute primary data that can inform novel discovery. Using a computationally intensive physics-based analytical platform operating at supercomputing speeds, we probed a high-resolution protein X-ray crystallographic library developed by us. For each of the eight identified novel 3D structures, we analyzed binding of sixty million compounds. Top-ranking compounds were acquired and screened for efficacy against breast, prostate, colon, or lung cancer, and for toxicity on normal human bone marrow stem cells, both using eight-day colony formation assays. Effective and non-toxic compounds segregated to two pockets. One compound, Dxr2-017, exhibited selective anti-melanoma activity in the NCI-60 cell line screen. In eight-day assays, Dxr2-017 had an IC50 of 12 nM against melanoma cells, while concentrations over 2100-fold higher had minimal stem cell toxicity. Dxr2-017 induced anoikis, a unique form of programmed cell death in need of targeted therapeutics. Our findings demonstrate proof-of-concept that protein structures represent high-value primary data to support the discovery of novel acting therapeutics. This approach is widely applicable.
Keywords: anoikis; computational biology; drug discovery; protein structure.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



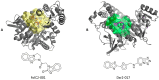
References
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

