A Comparison of the Cecal Microbiota between the Infection and Recovery Periods in Chickens with Different Susceptibilities to Eimeria tenella
- PMID: 39335298
- PMCID: PMC11428751
- DOI: 10.3390/ani14182709
A Comparison of the Cecal Microbiota between the Infection and Recovery Periods in Chickens with Different Susceptibilities to Eimeria tenella
Abstract
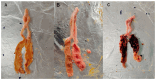
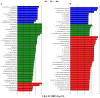
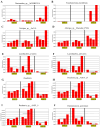
To investigate the effect of Eimeria tenella (E. tenella) infection on the cecal microbiota, resistant and susceptible families were screened out based on the coccidiosis resistance evaluation indexes after E. tenella infection. Subsequently, a comparative analysis of cecal microorganisms among control, resistant, and susceptible groups as well as between different periods following the E. tenella challenge was conducted using metagenomic sequencing technology. The results showed that the abundance of opportunistic pathogens, such as Pantoea, Sporomusa, and Pasteurella in the susceptible group and Helicobacter and Sutterella in the resistant group, was significantly higher on day 27 post-inoculation (PI) (the recovery period) than on day 5 PI (the infection period). Additionally, the abundance of Alistipes, Butyricicoccus, and Eubacterium in the susceptible group and Coprococcus, Roseburia, Butyricicoccus, and Lactobacillus in the resistant group showed a significant upward trend during the infection period compared with that in the recovery period. On day 5 PI, the abundance of Faecalibacterium and Lactobacillus was decreased in both the resistant and susceptible groups when compared with that in the control group and was greater in the resistant group than in the susceptible group, while Alistipes in the susceptible group had a relatively higher abundance than that in other groups. A total of 49 biomarker taxa were identified using the linear discriminant analysis (LDA) effect size (LEfSe) method. Of these, the relative abundance of Lactobacillus aviarius, Lactobacillus salivarius, Roseburia, and Ruminococcus gauvreauii was increased in the resistant group, while Bacteroides_sp__AGMB03916, Fusobacterium_mortiferum, Alistipes_sp__An31A, and Alistipes_sp__Marseille_P5061 were enriched in the susceptible group. On day 27 PI, LDA scores identified 43 biomarkers, among which the relative abundance of Elusimicrobium_sp__An273 and Desulfovibrio_sp__An276 was increased in the resistant group, while that of Bacteroides_sp__43_108, Chlamydiia, Chlamydiales, and Sutterella_sp__AM11 39 was augmented in the susceptible group. Our results indicated that E. tenella infection affects the structure of the cecal microbiota during both the challenge and recovery periods. These findings will enhance the understanding of the effects of changes in the cecal microbiota on chickens after coccidia infection and provide a reference for further research on the mechanisms underlying how the intestinal microbiota influence the growth and health of chickens.
Keywords: E. tenella; cecal microbiota; metagenome sequencing; resistance selection.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures






References
-
- Shirley M.W., Smith A.L., Tomley F.M. The biology of avian Eimeria with an emphasis on their control by vaccination. Adv. Parasitol. 2005;60:285–330. - PubMed
-
- Li X., Jiang X.J., Qi D.X., Wang X., Wang C., Fei C., Zhou W., Li J., Zhang K. Effects of ethanamizuril, sulfachlorpyridazine or their combination on cecum microbial community and metabolomics in chickens infected with Eimeria tenella. Microb. Pathog. 2022;173:105823. doi: 10.1016/j.micpath.2022.105823. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

