Genomic Regions Associated with Growth and Reproduction Traits in Pink-Eyed White Mink
- PMID: 39336733
- PMCID: PMC11431770
- DOI: 10.3390/genes15091142
Genomic Regions Associated with Growth and Reproduction Traits in Pink-Eyed White Mink
Abstract
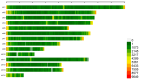
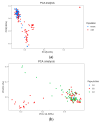
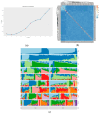
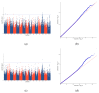
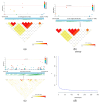
In mink breeding, balanced selection for growth and reproductive features is essential because these traits are contradictory. The variables of total number born (TNB), number born alive (NBA), and body weight (BW) are highly valuable in terms of their importance in mink production. A comprehensive understanding of the molecular mechanisms that drive these features could offer vital insights into their genetic compositions. In the present study, the single-nucleotide polymorphism (SNP) genotypes of 219 minks were obtained via double digest restriction-site associated DNA sequencing (ddRAD-seq). Following several rounds of screening, about 2,415,121 high-quality SNPs were selected for a genome-wide association study (GWAS). The GWAS was used to determine BW and reproductive traits in pink-eyed white mink. It was suggested that SLC26A36, STXBP5L, and RPS 29 serve as potential genes for the total number of kits born (TNB), while FSCB, PDPN, NKX 2-1, NFKB 1, NFKBIA, and GABBR1 are key genes for the number born alive (NBA). Moreover, RTTN, PRPF31, MACROD1, and KYAT1 are possible BW genes based on association results and available functional data from gene and mammalian phenotype databases. These results offer essential information about the variety of mink and theoretical principles for applying mink breeds.
Keywords: body weight; genome-wide association study; mink; number born alive; single-nucleotide polymorphism; total number born.
Conflict of interest statement
The author declare no conflict of interest.
Figures

References
-
- Nes N., Einarsson E.J., Lohi O., Jorgensen G. Beautiful Fur Animals and Their Colour Genetics. Scientific; Copenhagen, Denmark: 1988.
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

