Uncovering a Genetic Diagnosis in a Pediatric Patient by Whole Exome Sequencing: A Modeling Investigation in Wiedemann-Steiner Syndrome
- PMID: 39336746
- PMCID: PMC11431573
- DOI: 10.3390/genes15091155
Uncovering a Genetic Diagnosis in a Pediatric Patient by Whole Exome Sequencing: A Modeling Investigation in Wiedemann-Steiner Syndrome
Abstract
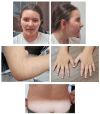
Background: Wiedemann-Steiner syndrome (WSS), a rare autosomal-dominant disorder caused by haploinsufficiency of the KMT2A gene product, is part of a group of disorders called chromatinopathies. Chromatinopathies are neurodevelopmental disorders caused by mutations affecting the proteins responsible for chromatin remodeling and transcriptional regulation. The resulting gene expression dysregulation mediates the onset of a series of clinical features such as developmental delay, intellectual disability, facial dysmorphism, and behavioral disorders. Aim of the Study: The aim of this study was to investigate a 10-year-old girl who presented with clinical features suggestive of WSS. Methods: Clinical and genetic investigations were performed. Whole exome sequencing (WES) was used for genetic testing, performed using Illumina technology. The bidirectional capillary Sanger resequencing technique was used in accordance with standard methodology to validate a mutation discovered by WES in all family members who were available. Utilizing computational protein modeling for structural and functional studies as well as in silico pathogenicity prediction models, the effect of the mutation was examined. Results: WES identified a de novo heterozygous missense variant in the KMT2A gene KMT2A(NM_001197104.2): c.3451C>G, p.(Arg1151Gly), absent in the gnomAD database. The variant was classified as Likely Pathogenetic (LP) according to the ACMG criteria and was predicted to affect the CXXC-type zinc finger domain functionality of the protein. Modeling of the resulting protein structure suggested that this variant changes the protein flexibility due to a variation in the Gibbs free energy and in the vibrational entropy energy difference between the wild-type and mutated domain, resulting in an alteration of the DNA binding affinity. Conclusions: A novel and de novo mutation discovered by the NGS approach, enhancing the mutation spectrum in the KMT2A gene, was characterized and associated with WSS. This novel KMT2A gene variant is suggested to modify the CXXC-type zinc finger domain functionality by affecting protein flexibility and DNA binding.
Keywords: KMT2A gene; Wiedemann–Steiner syndrome; modeling; whole exome sequencing.
Conflict of interest statement
The authors disclose that the research was conducted without any financial or commercial ties that could be perceived as conflicts of interest.
Figures




References
-
- Stenson P.D., Mort M., Ball E.V., Evans K., Hayden M., Heywood S., Hussain M., Phillips A.D., Cooper D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017;136:665–677. doi: 10.1007/s00439-017-1779-6. - DOI - PMC - PubMed
-
- Di Fede E., Massa V., Augello B., Squeo G., Scarano E., Perri A.M., Fischetto R., Causio F.A., Zampino G., Piccione M., et al. Expanding the phenotype associated to KMT2A variants: Overlapping clinical signs between Wiedemann-Steiner and Rubinstein-Taybi syndromes. Eur. J. Hum. Genet. EJHG. 2021;29:88–98. doi: 10.1038/s41431-020-0679-8. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

